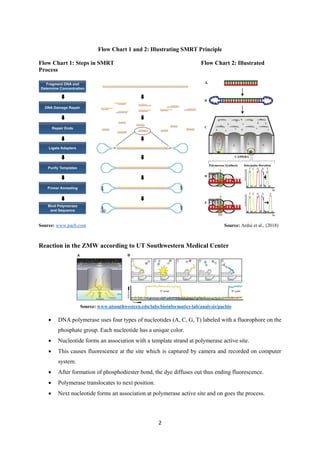
Single Molecule Real-Time (SMRT) sequencing developed by Pacific Biosciences is a third-generation long-read technology that enables accurate DNA sequencing using a polymerase enzyme to detect fluorescently labeled nucleotides in real-time. It has applications in whole genome sequencing, RNA sequencing, and epigenetic studies, providing advantages such as long read lengths, uniform coverage, and high accuracy, but also has disadvantages like high costs, sequencing error rates, and lower throughput compared to other technologies. Overall, SMRT sequencing offers powerful capabilities for genomic research despite its limitations.