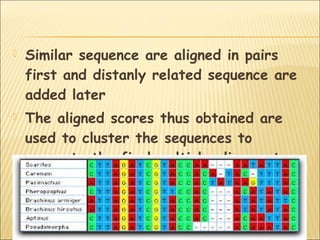
Sequence alignment involves arranging two or more biological sequences, like DNA, RNA or proteins, in a way that optimally matches their elements. It helps infer functional, structural or evolutionary relationships between sequences. There are two main types of sequence alignment - global alignment, which aligns entire sequences end-to-end, and local alignment, which finds short, locally matching regions. Popular algorithms for sequence alignment include BLAST, FASTA and CLUSTAL, with BLAST being the most widely used due to its speed.