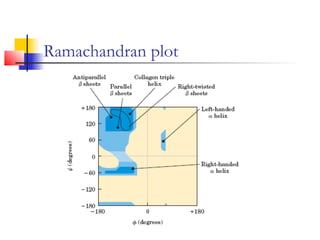
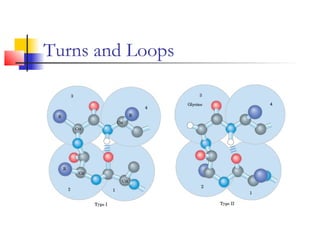
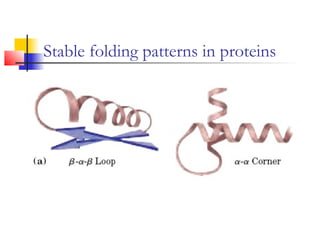
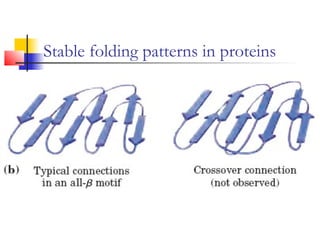
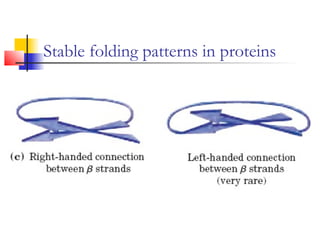
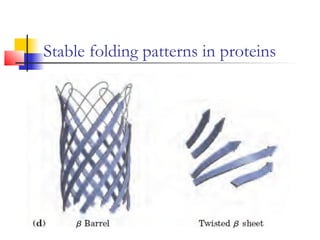
This document discusses protein structure, classification, prediction, and visualization. It covers secondary structure elements like alpha helices and beta sheets, as well as tertiary and quaternary structure. It describes protein structure databases like the Protein Data Bank and tools for visualizing protein structures. Different amino acid properties that influence secondary structure are also discussed.