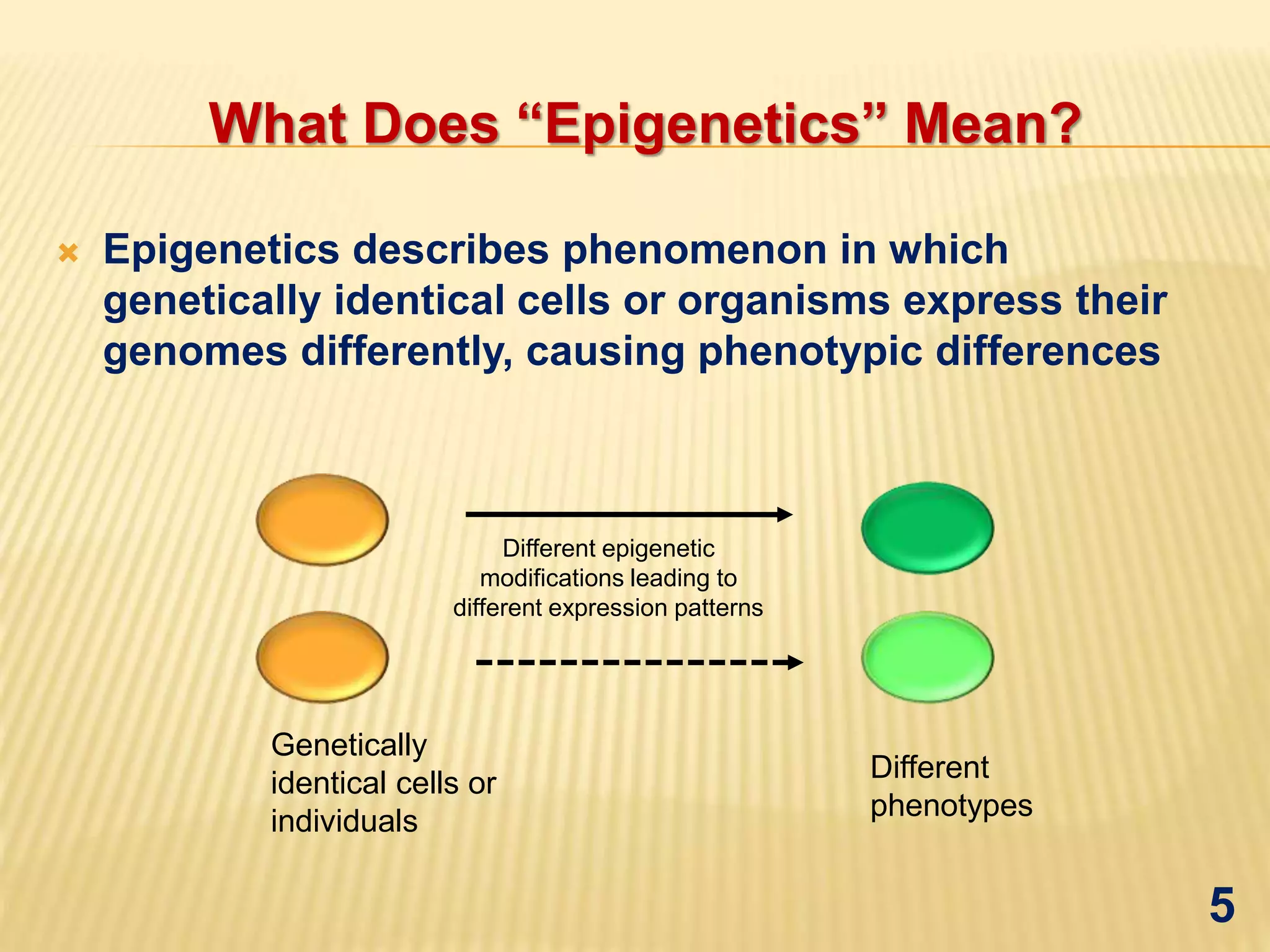
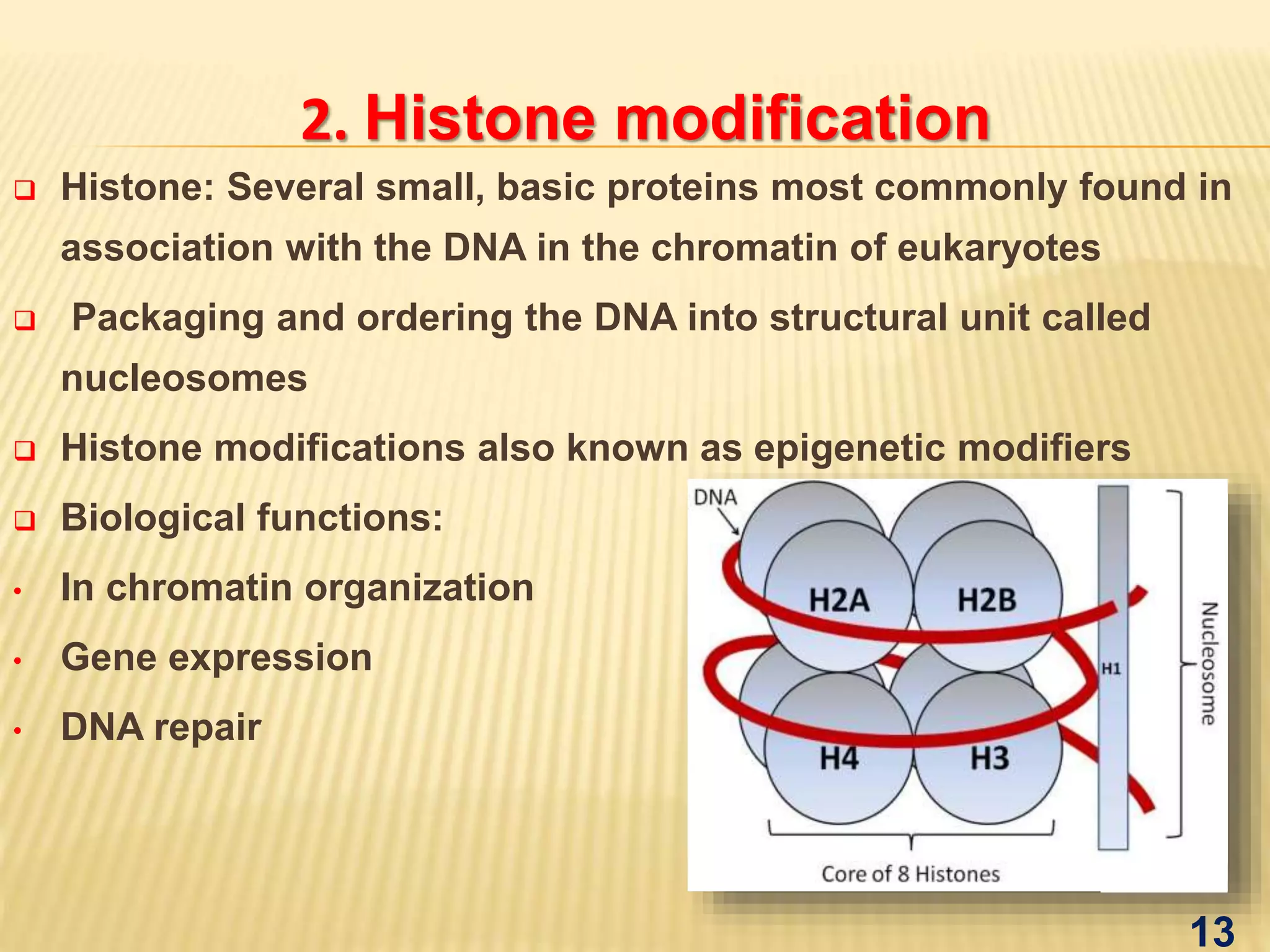
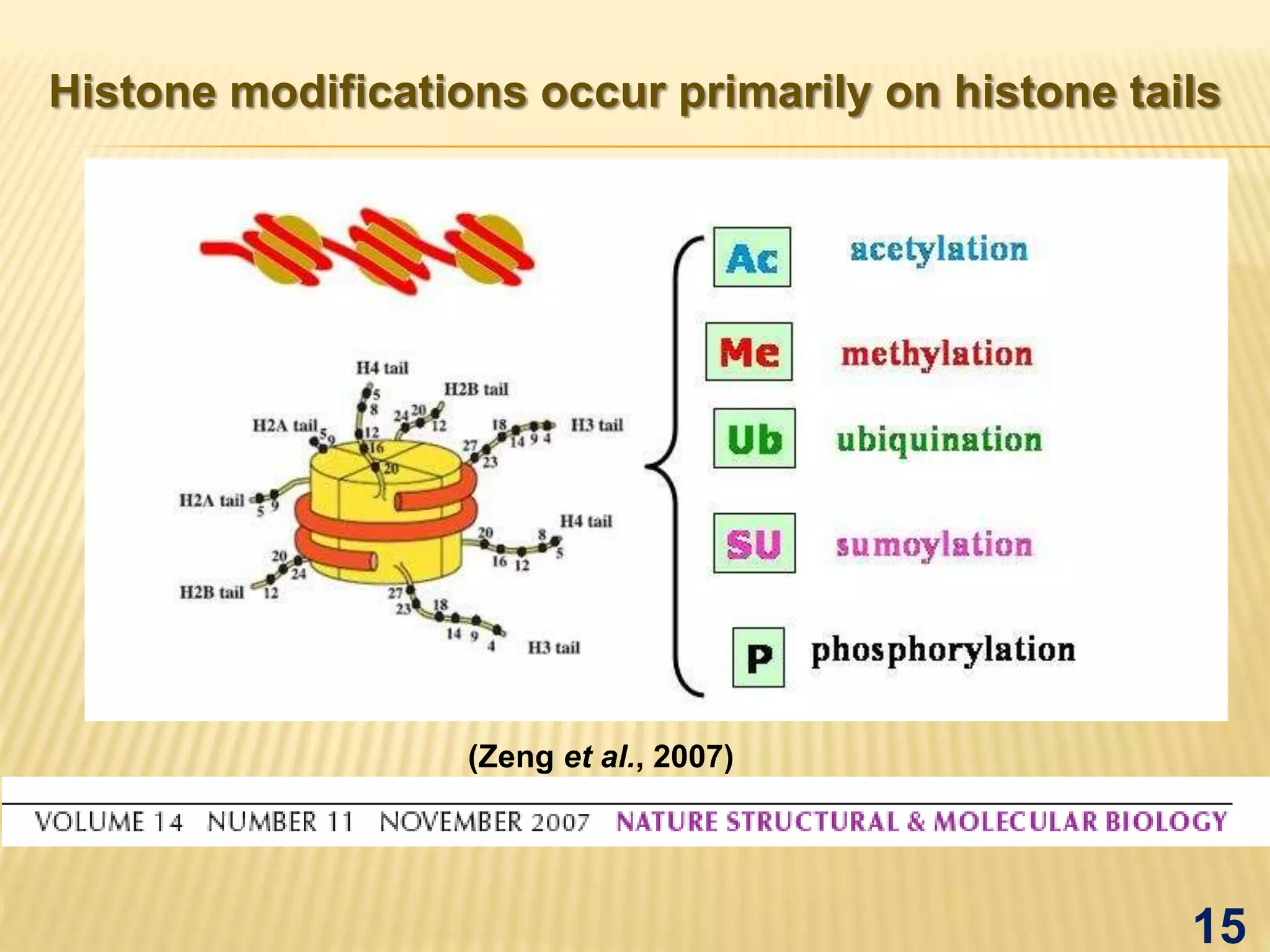
Epigenetics mediated gene regulation in plants describes how epigenetic mechanisms like DNA methylation and histone modification can regulate gene expression without changing the underlying DNA sequence. The document discusses various epigenetic mechanisms in plants including DNA methylation, histone modifications, and RNA interference. It also outlines applications of epigenetics like improving plant stress tolerance and yield as well as evolutionary studies.