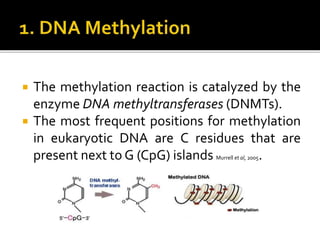
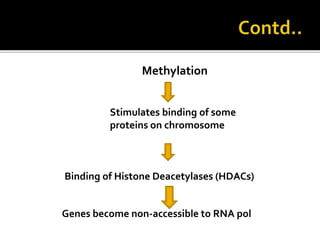
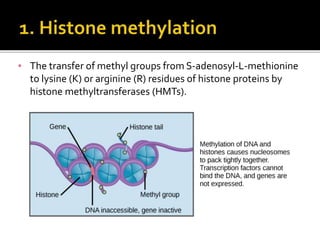
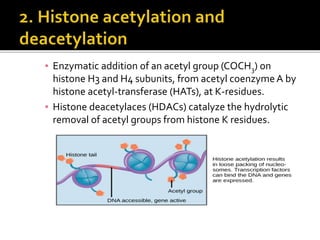
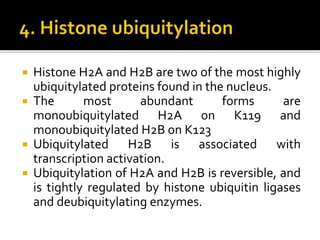
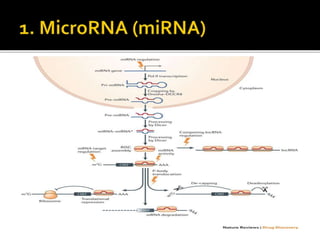
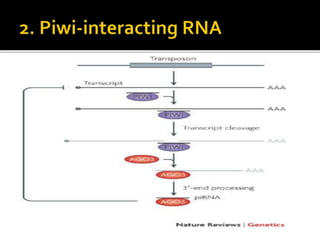
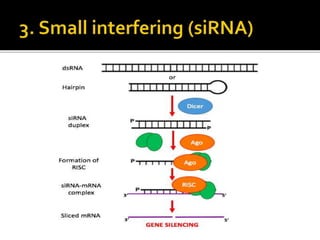
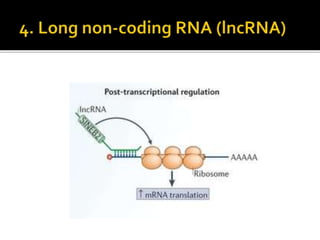
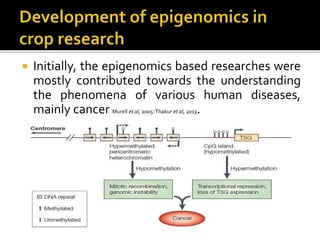
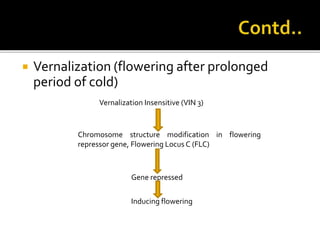
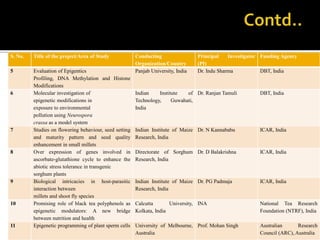
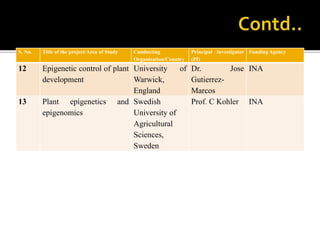
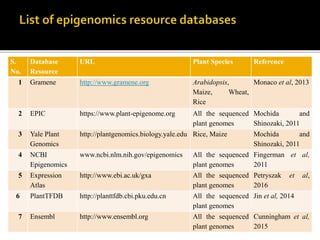
The document discusses epigenetics and epigenomics in plants, describing the main epigenetic modifications including DNA methylation, histone modifications, and non-coding RNAs. It reviews several ongoing research projects applying epigenomics to improve crop traits and stress resistance in important crops like wheat, rice, and maize. Going forward, further research is needed to better understand how epigenetic changes influence plant development and physiology, their degree of heritability, and how epigenomics can be used to enhance crop breeding.