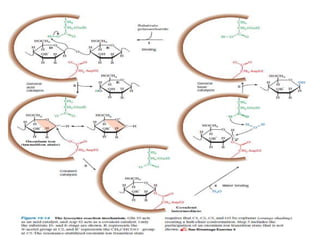
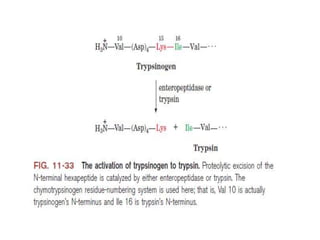
1. Lysozyme attaches to bacterial cell walls and uses its Glu 35 residue to transfer a proton to cleave the bond between the D- and E-rings of the cell wall, forming an oxonium ion transition state.
2. The Asp 52 carboxylate group then nucleophilically attacks the electron-poor carbon of the D-ring to form a glycosyl-enzyme intermediate.
3. Hydrolysis of this intermediate by Glu 35, forming another transition state, completes the catalytic cycle and releases the D-ring product.