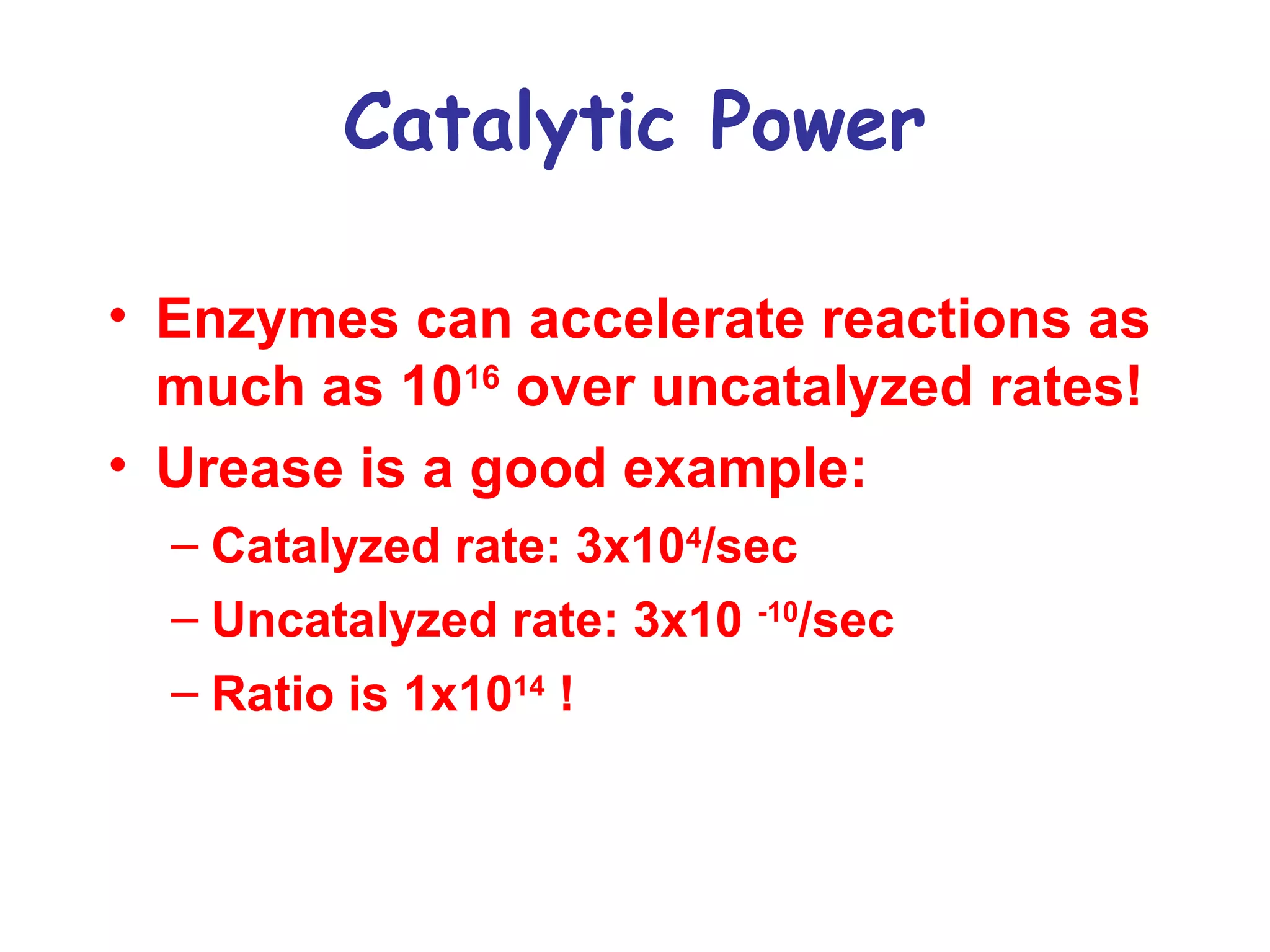
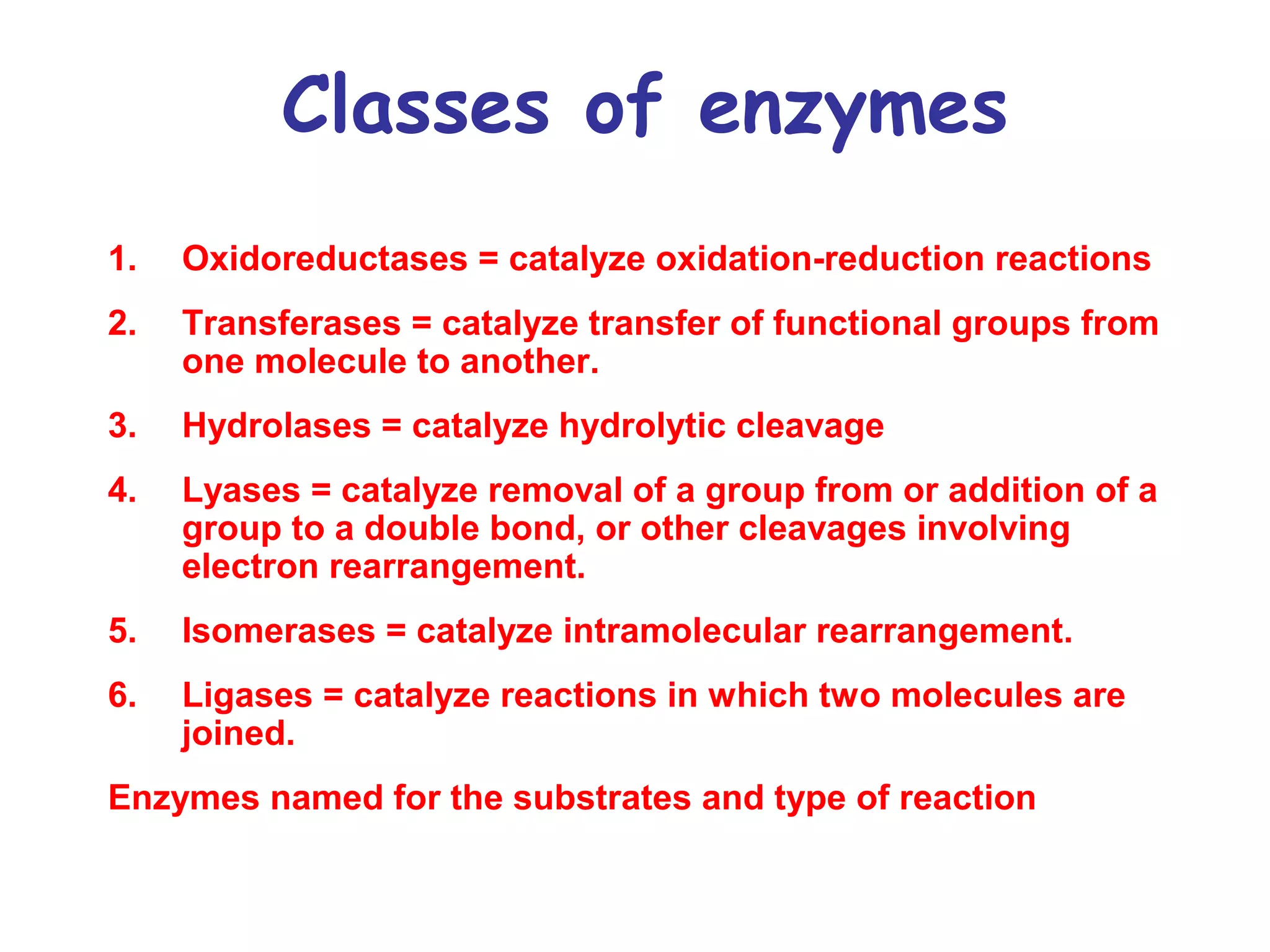
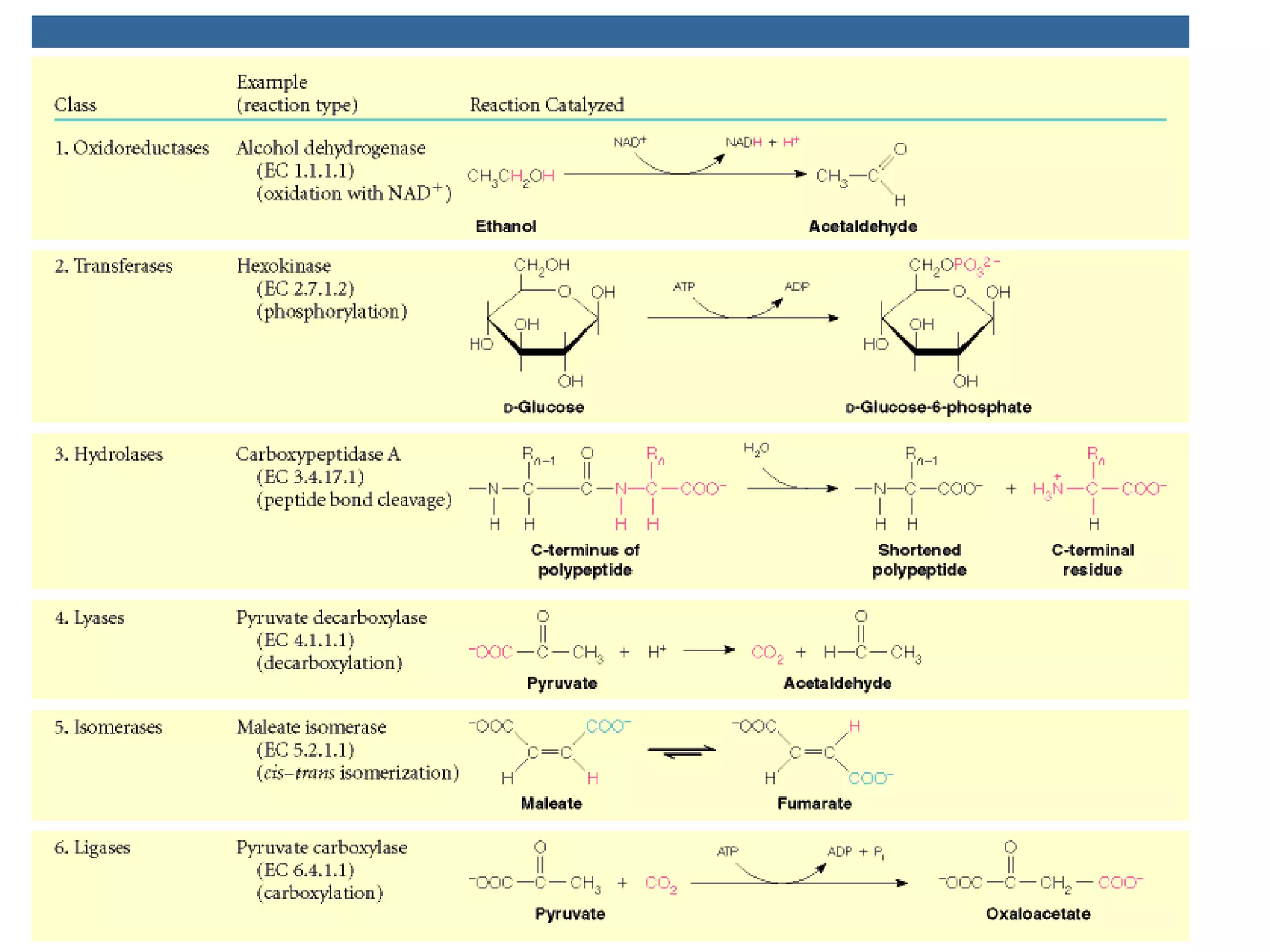
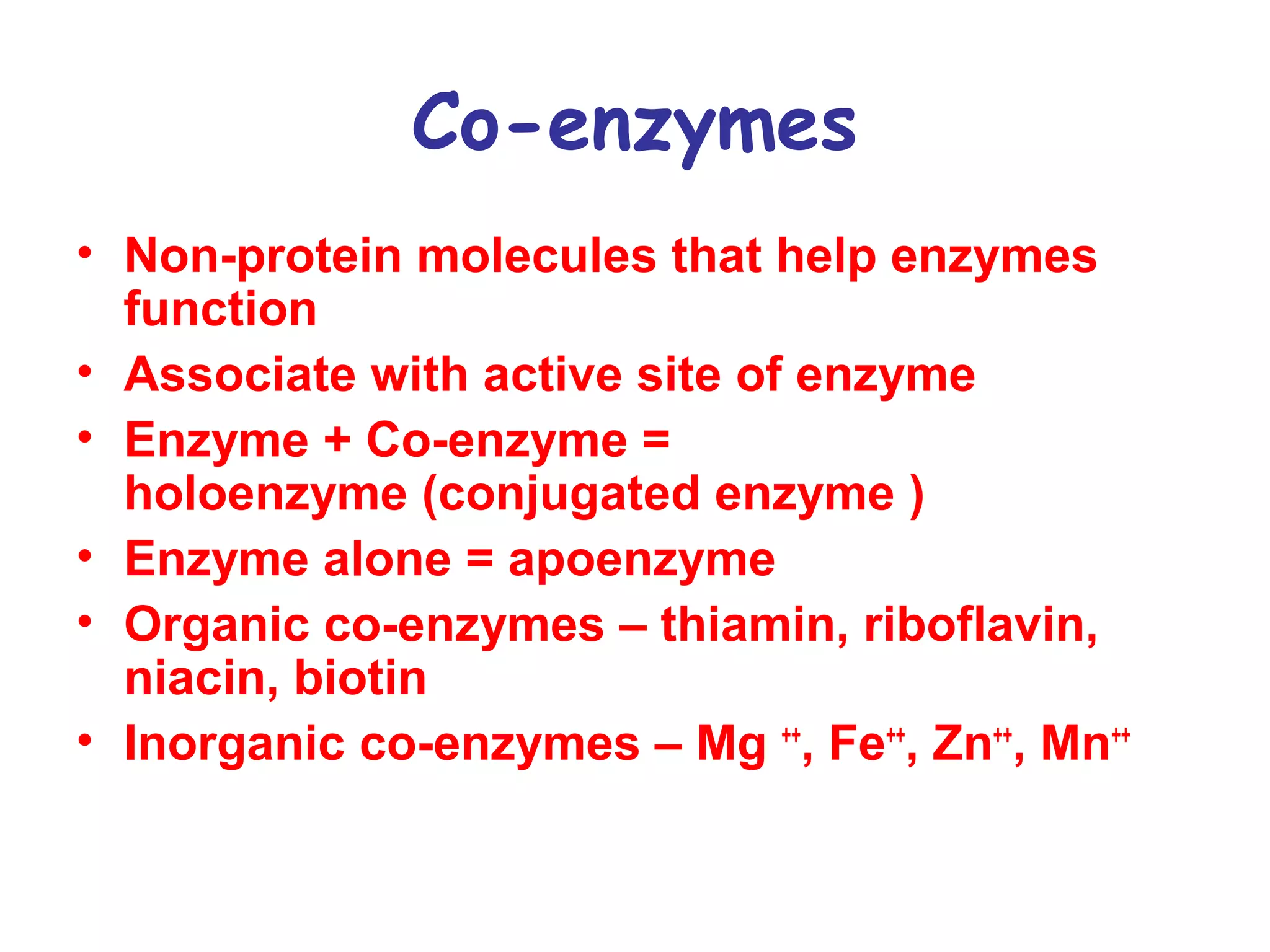
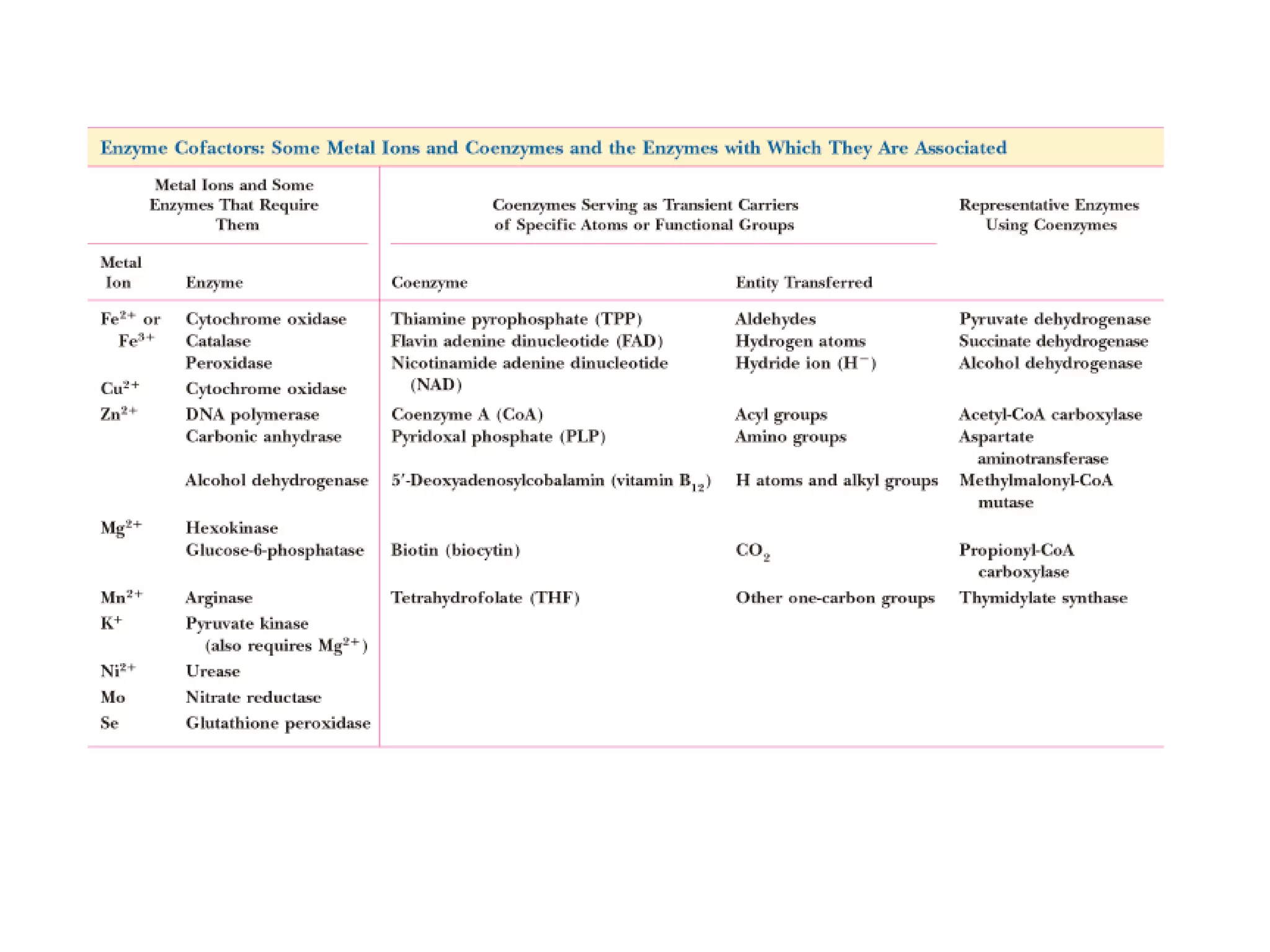
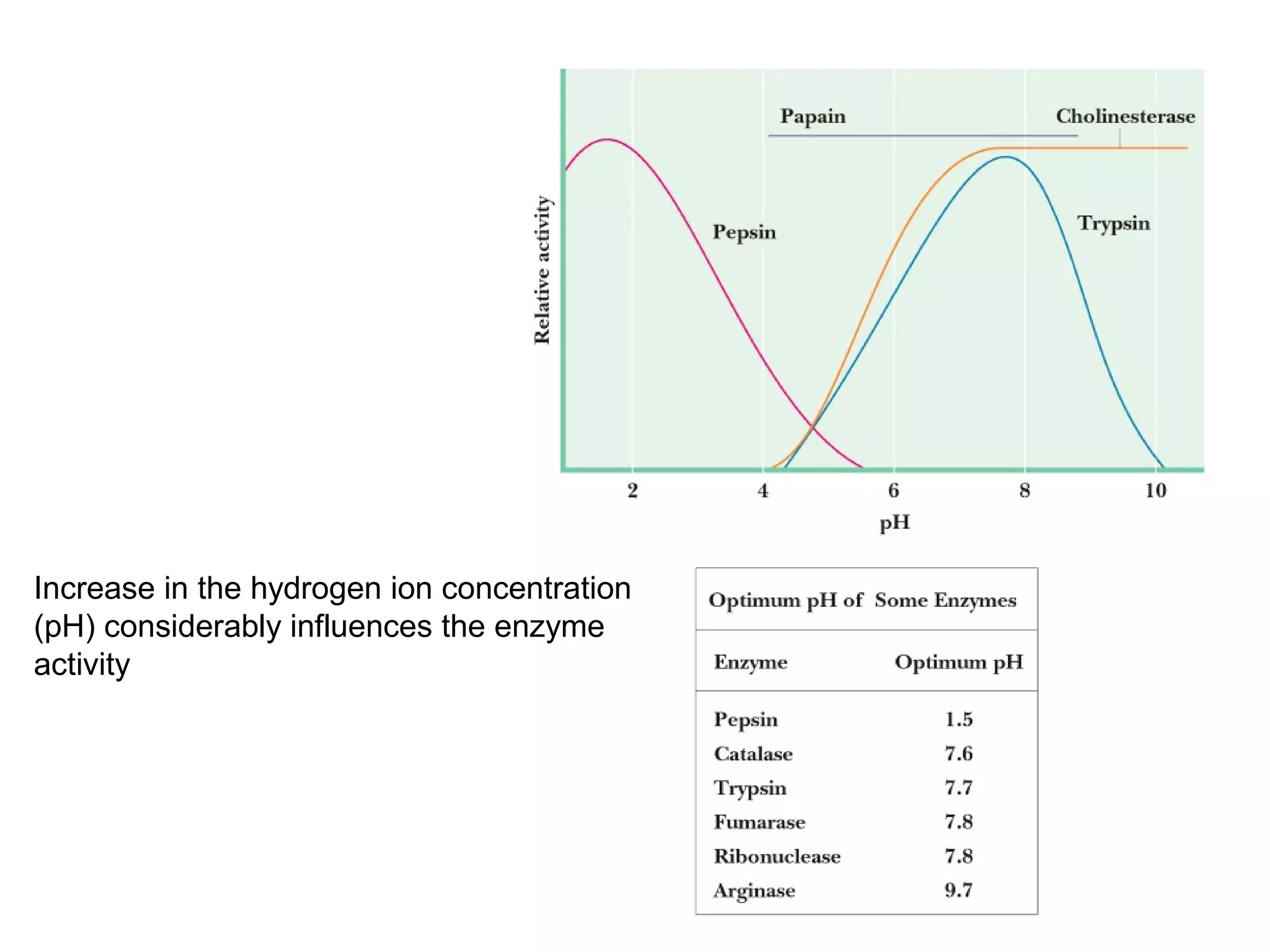
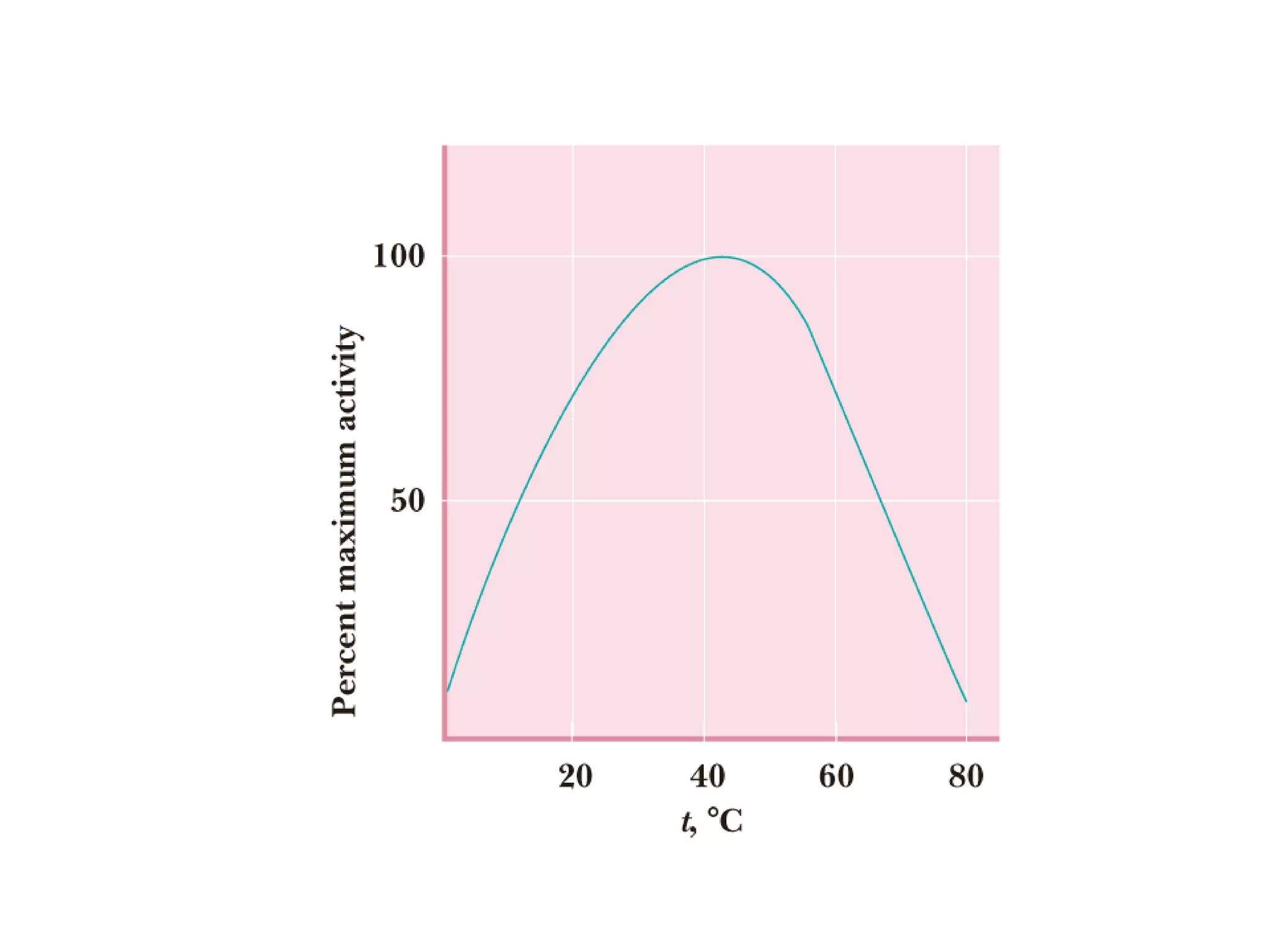
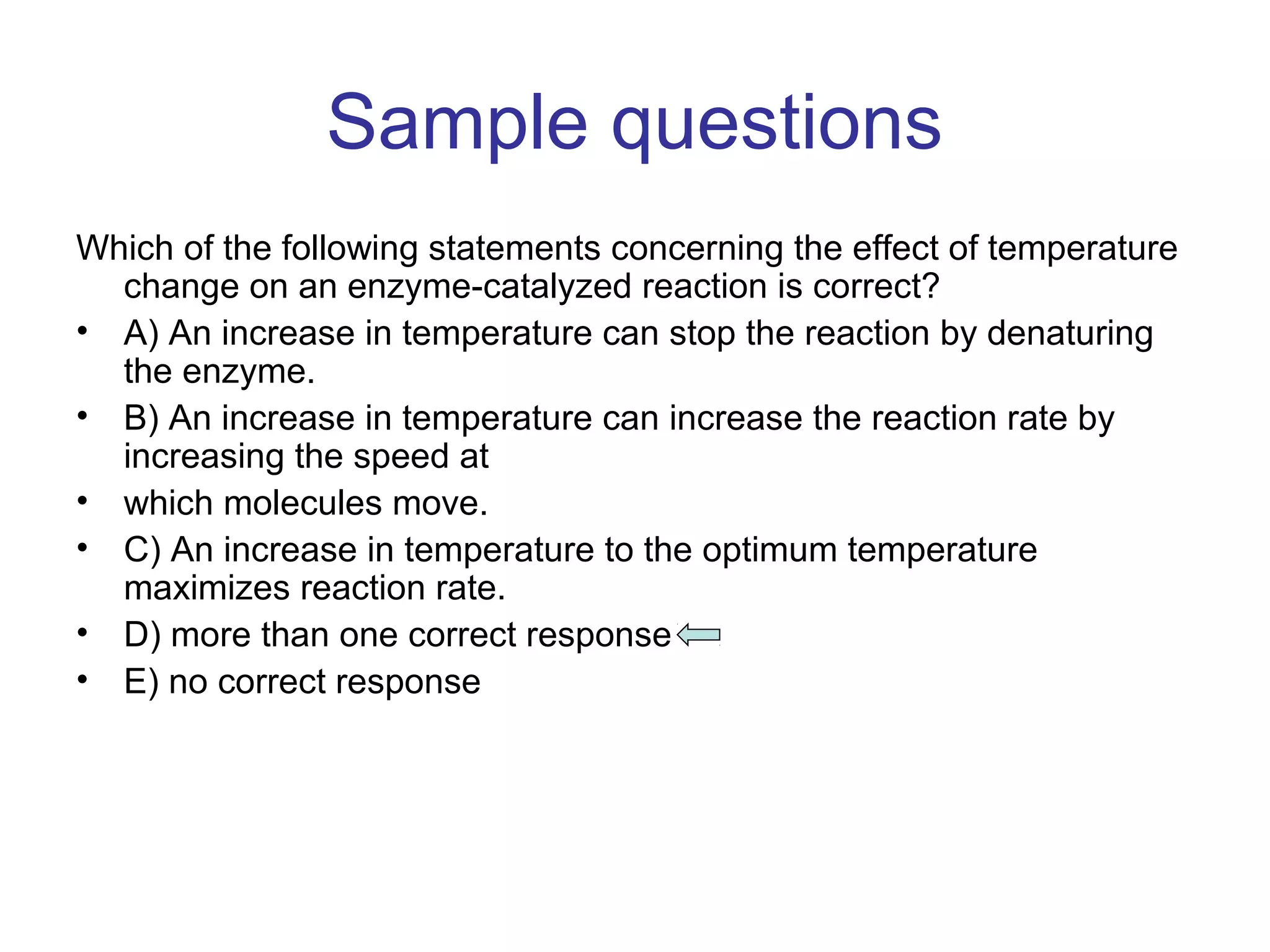
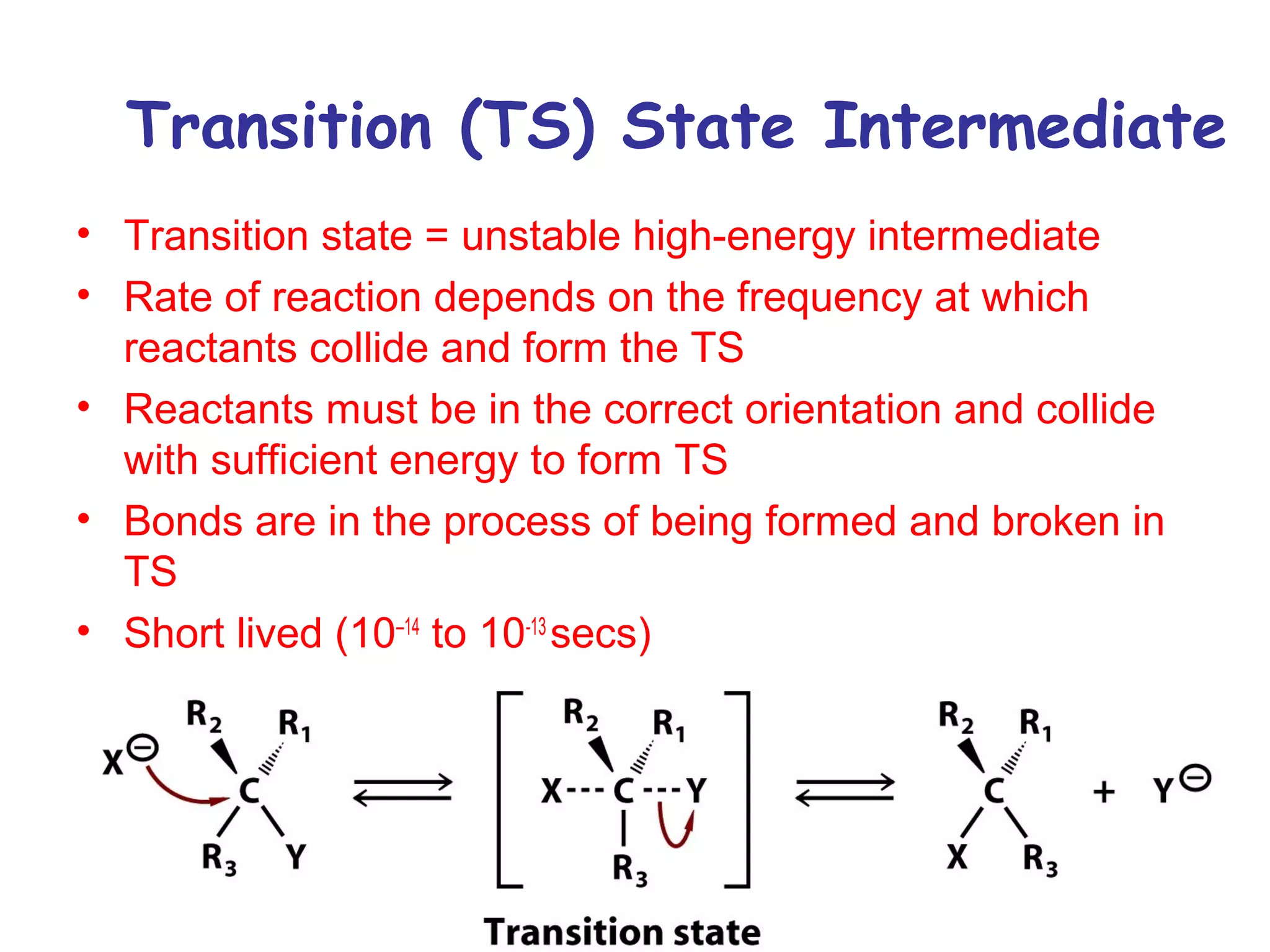
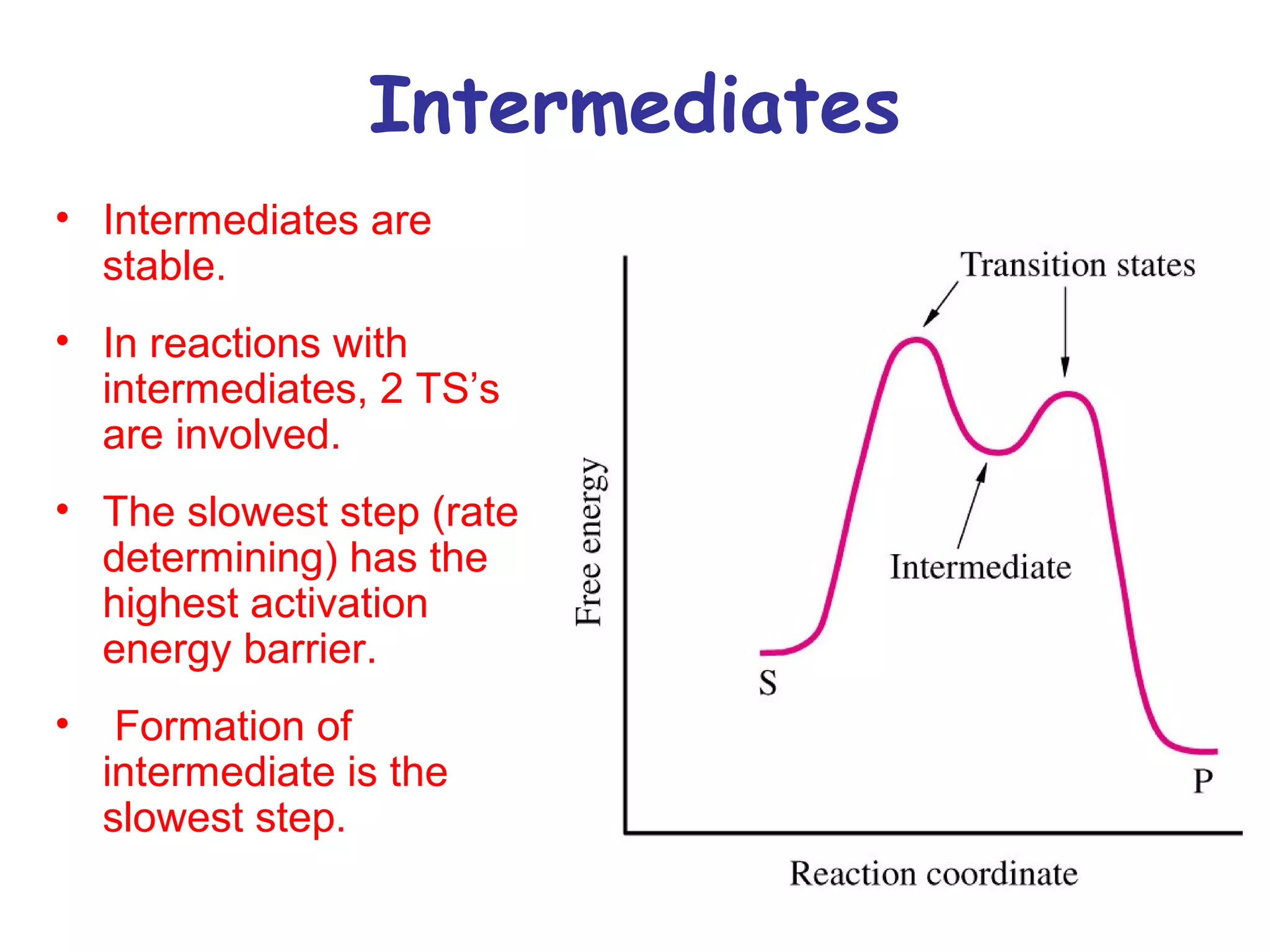
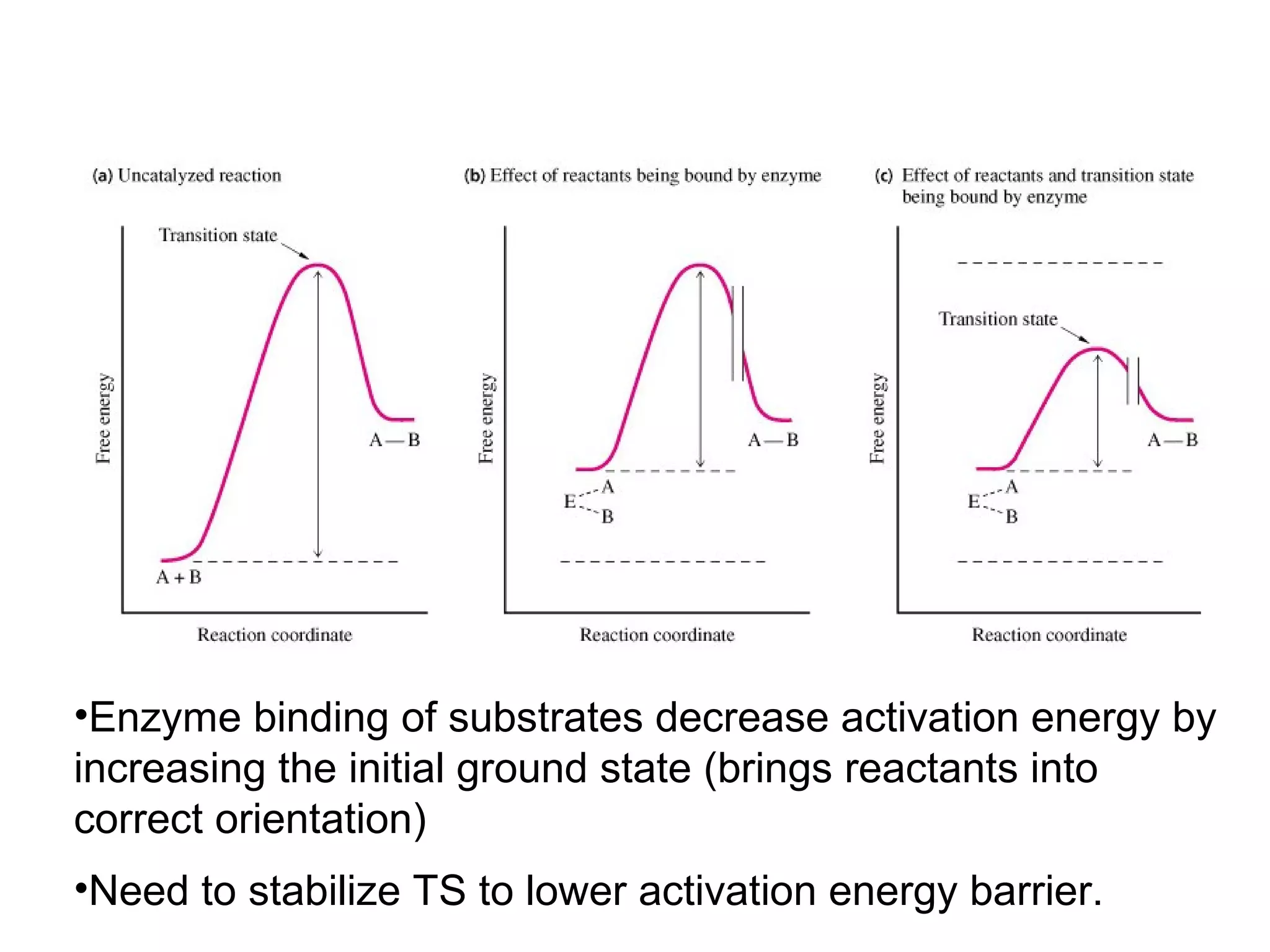
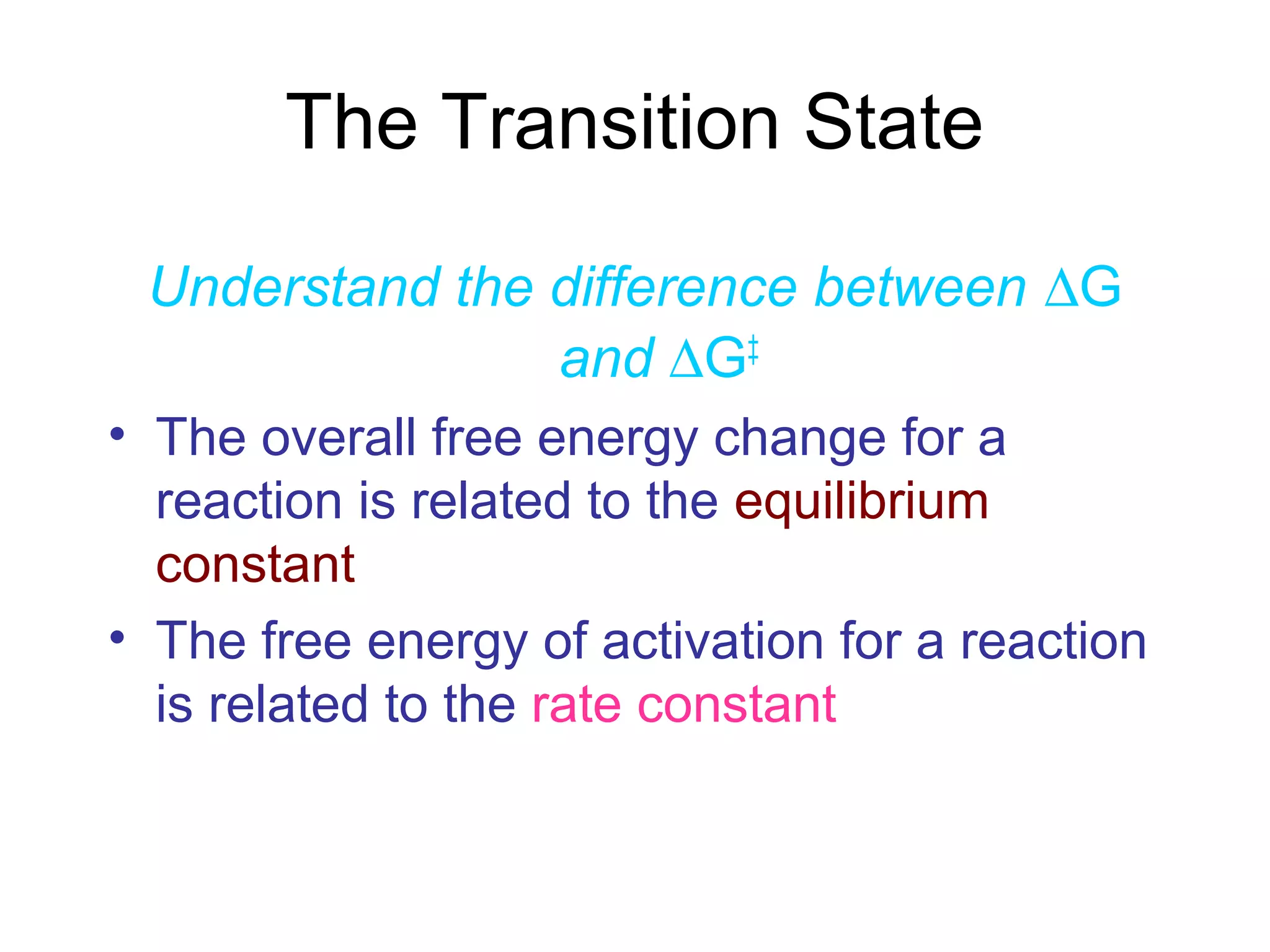
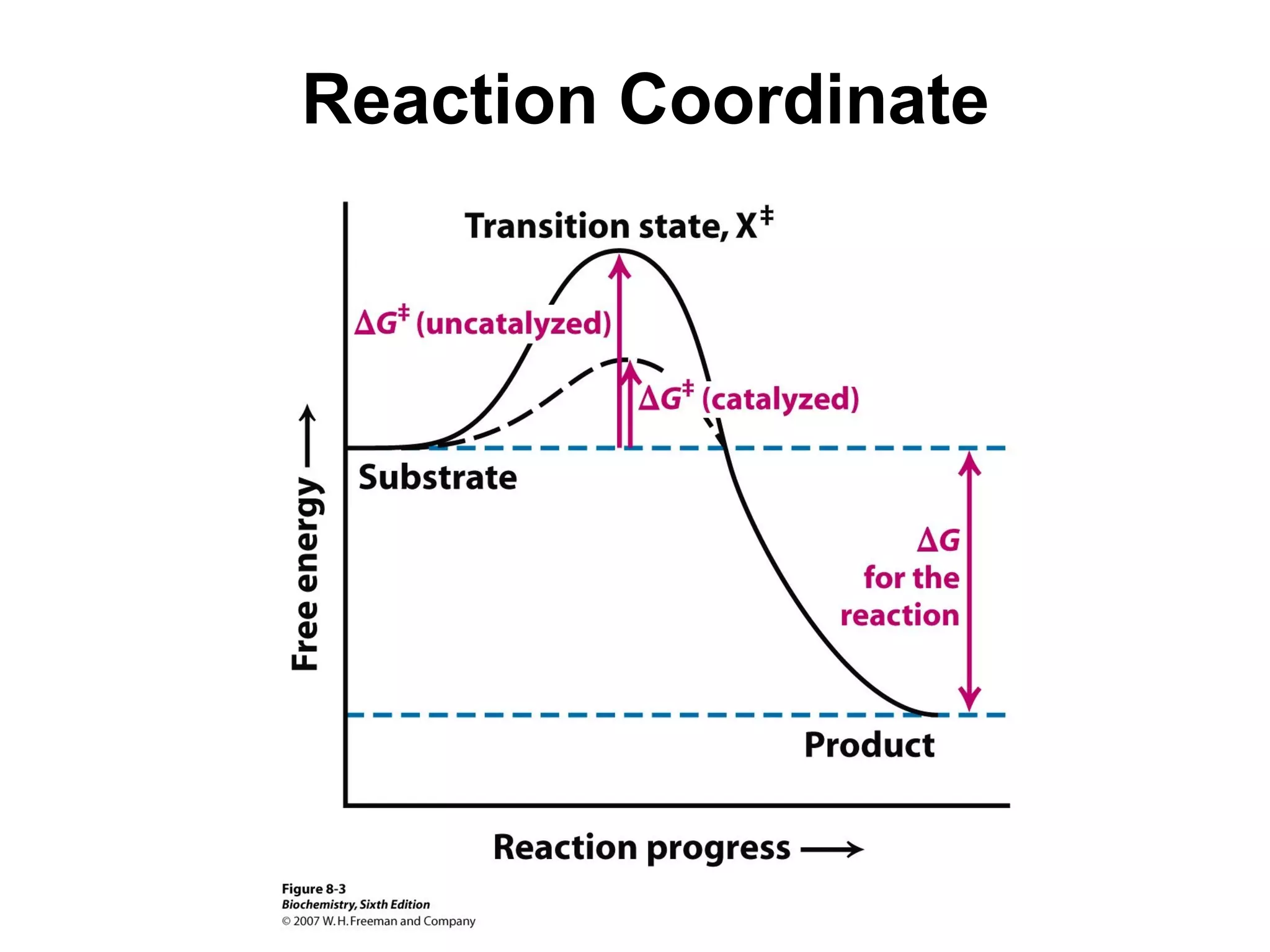
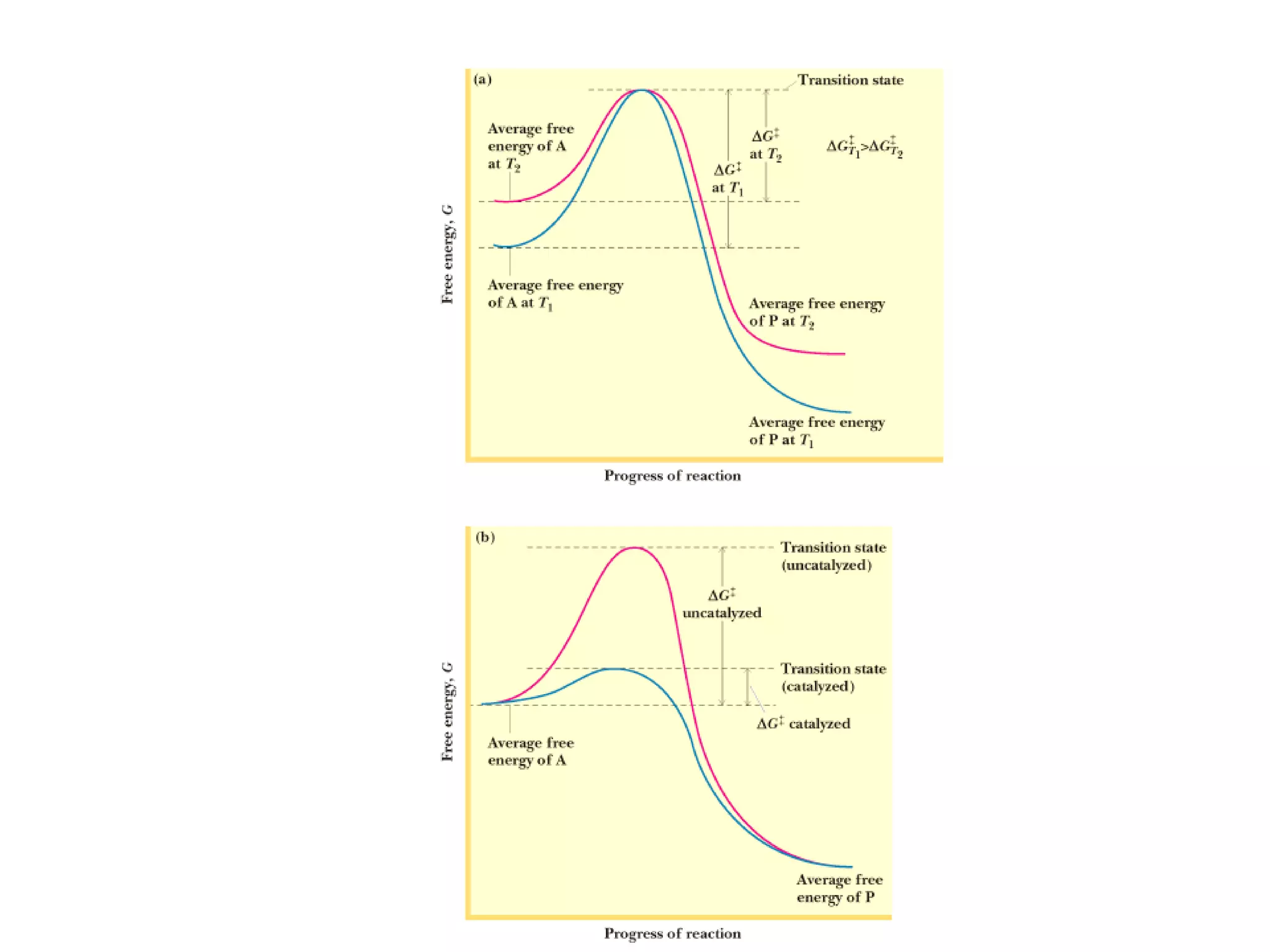
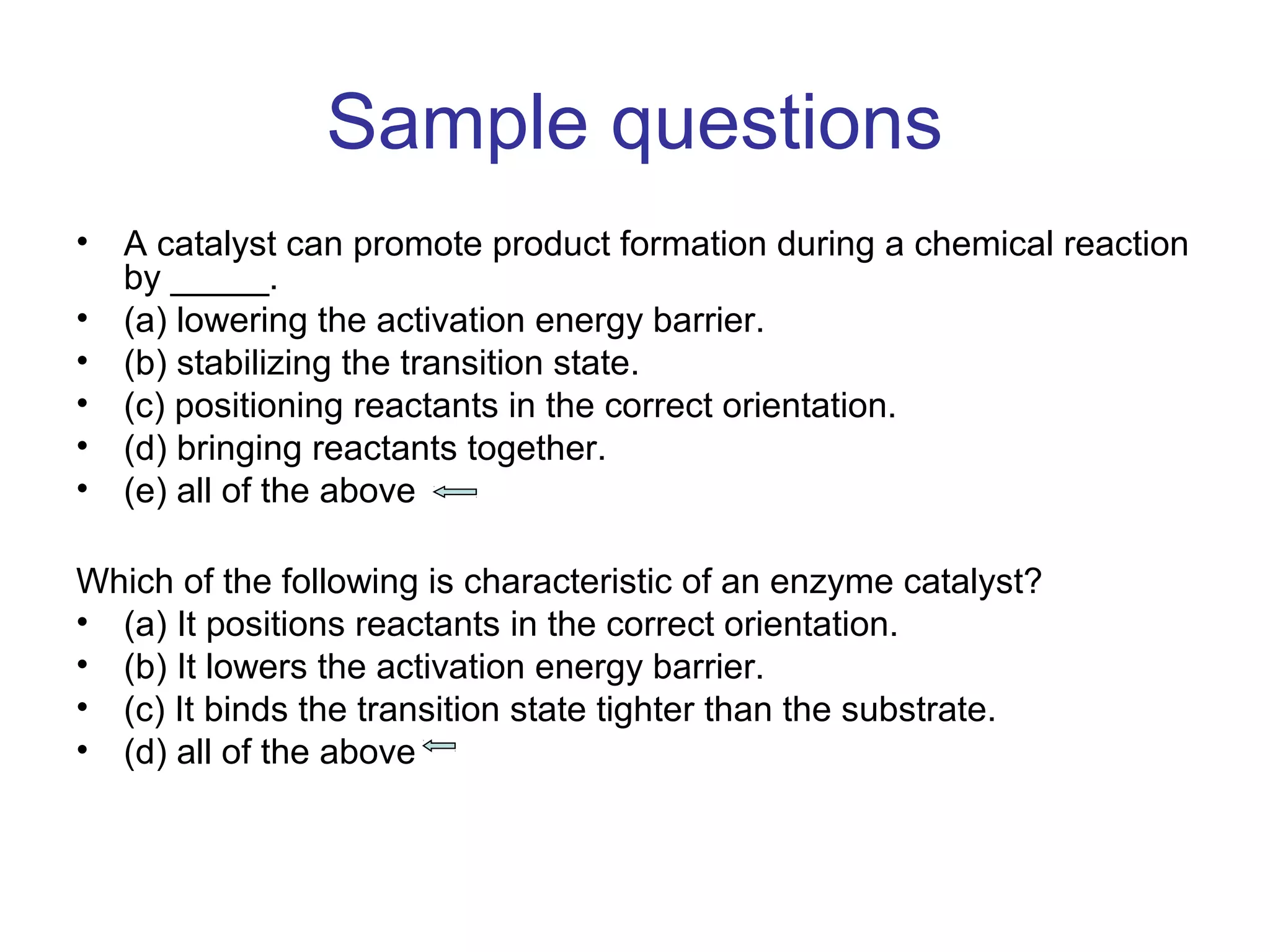
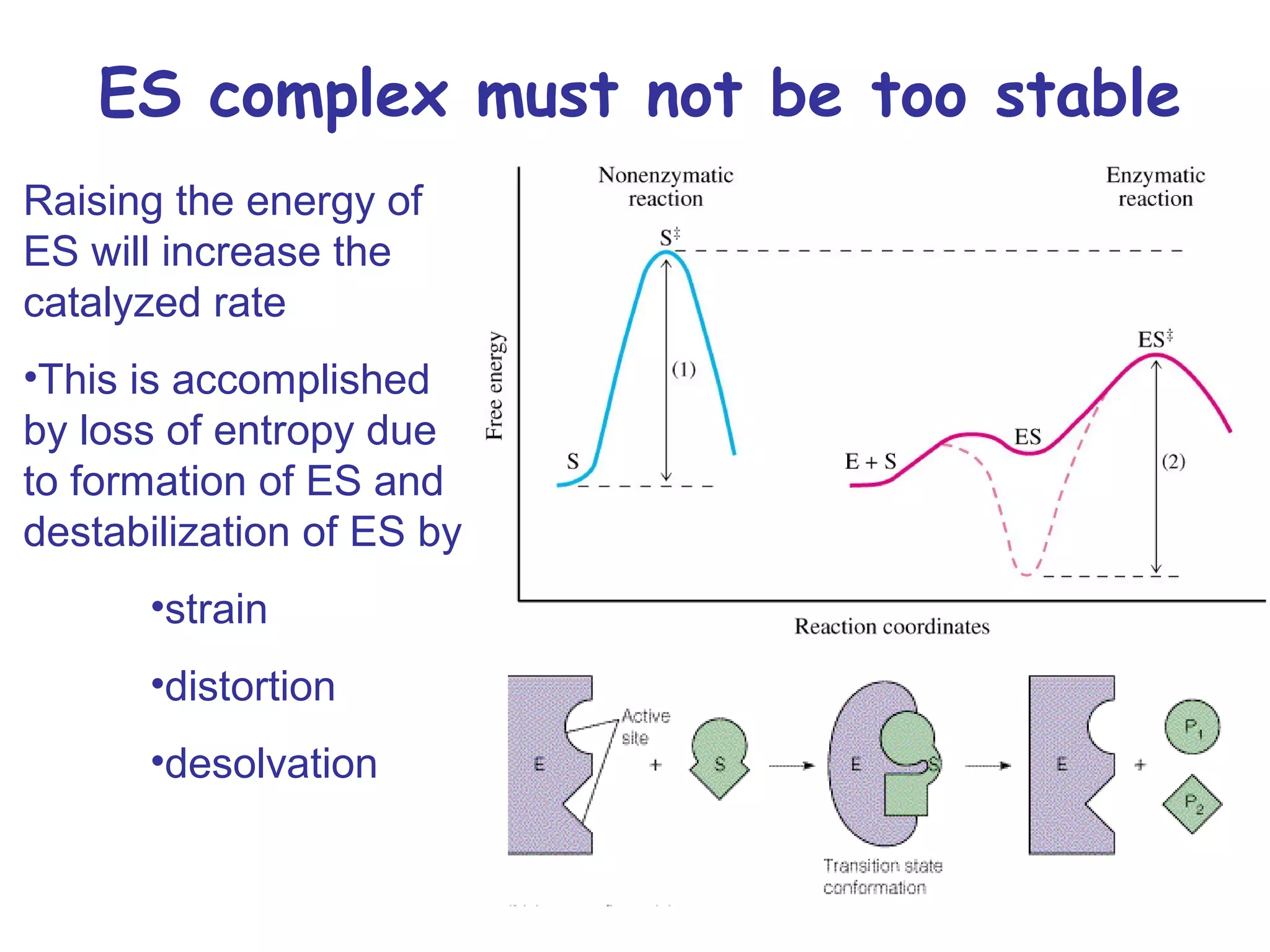
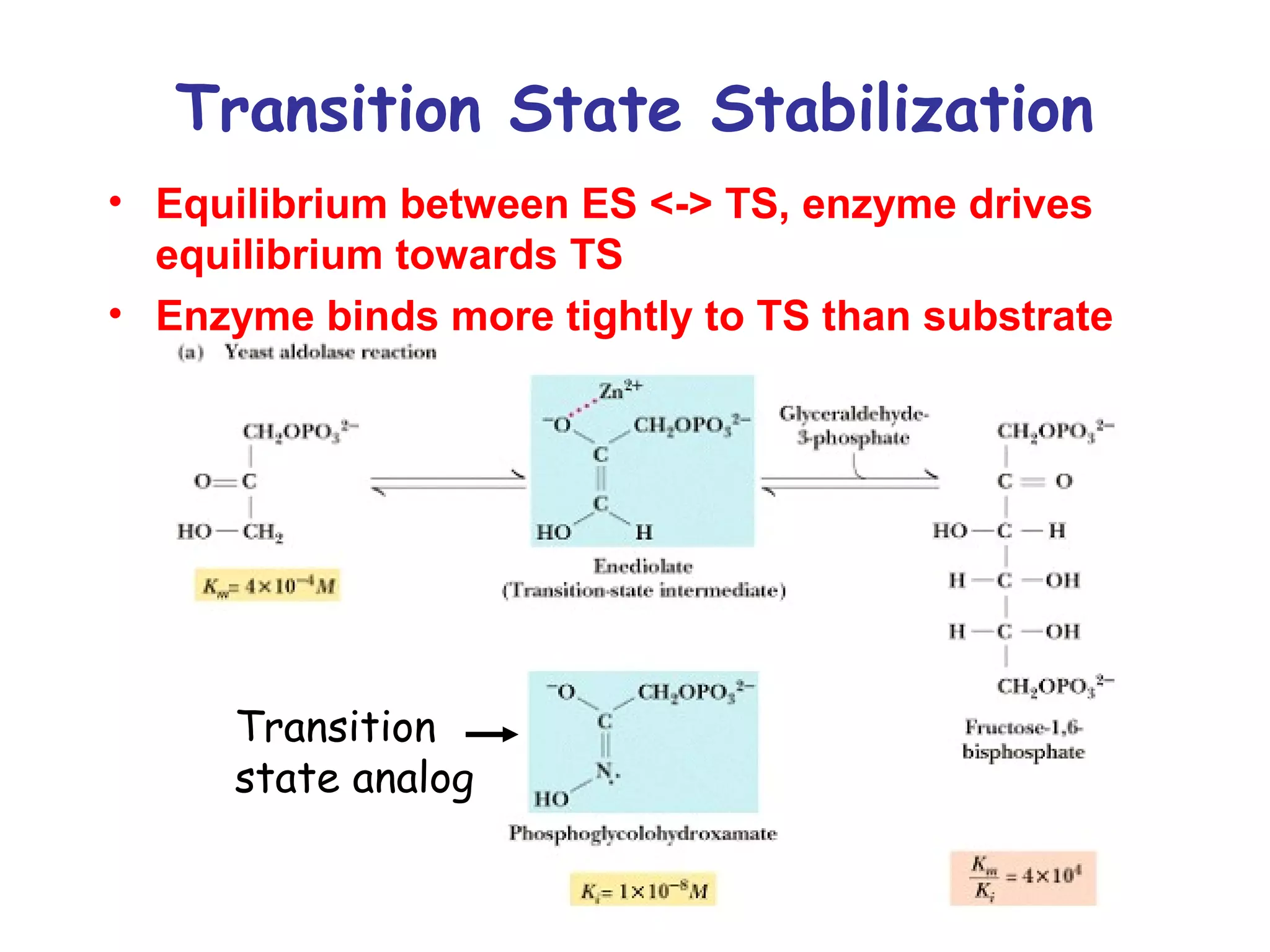
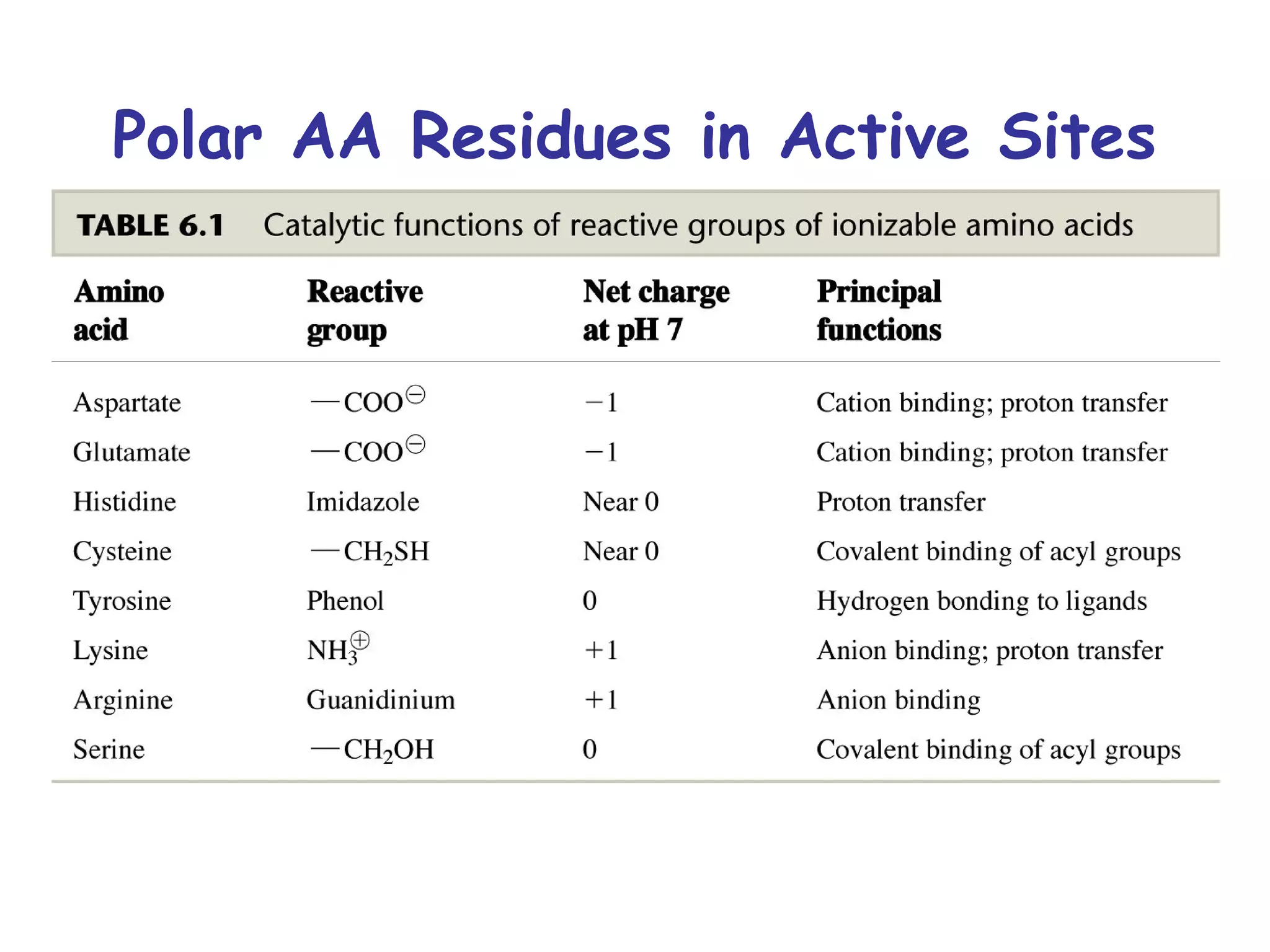
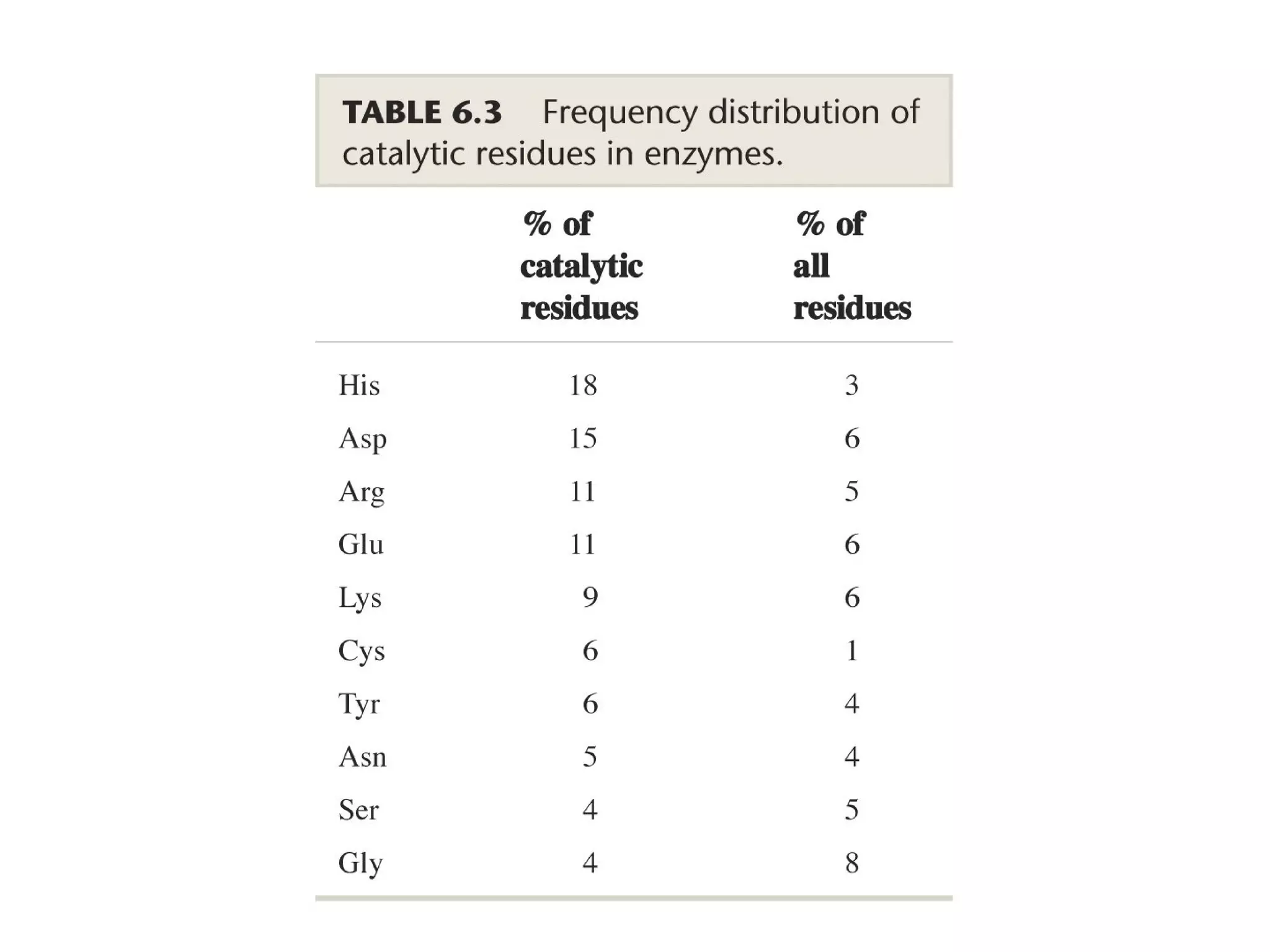
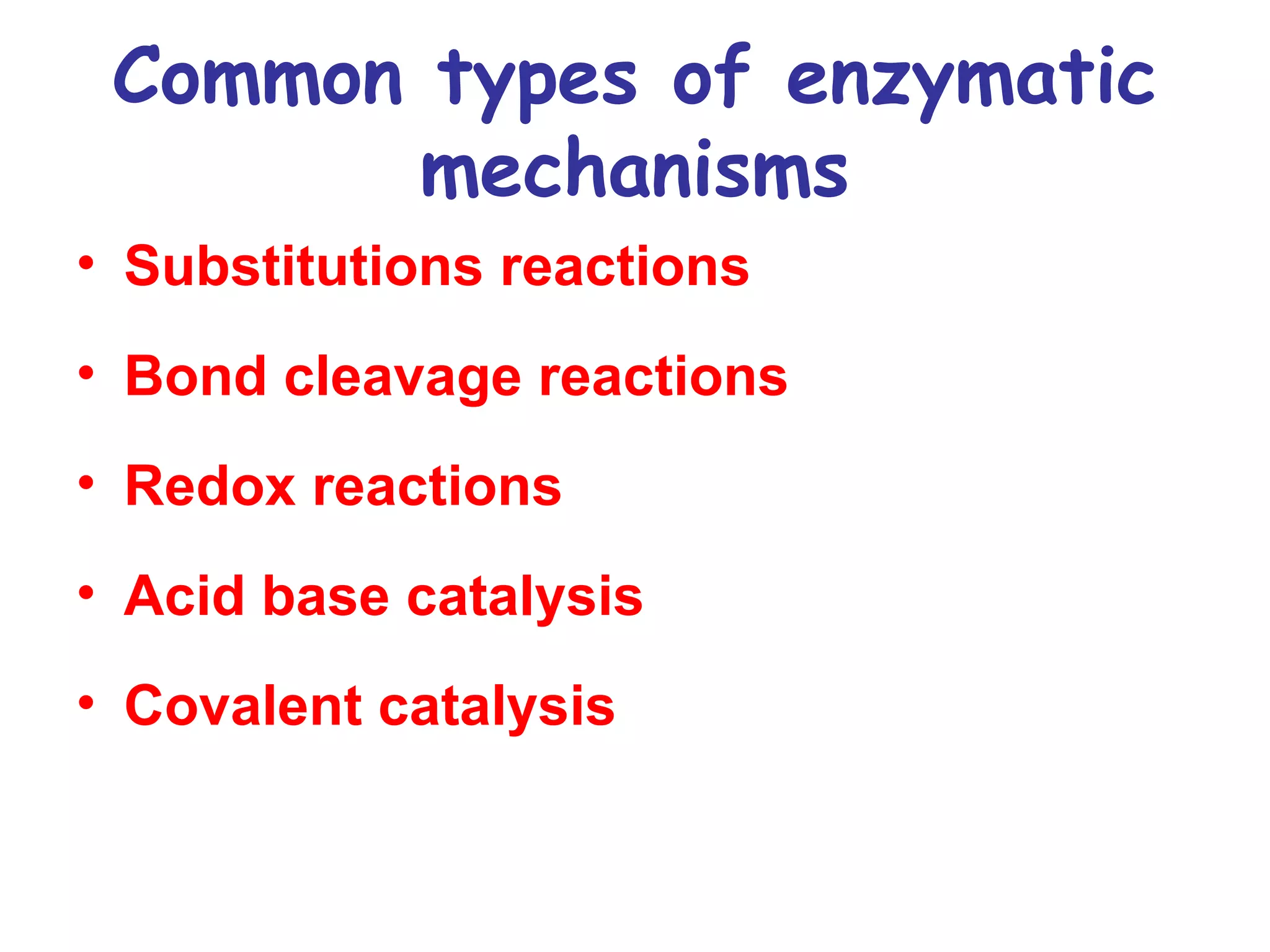
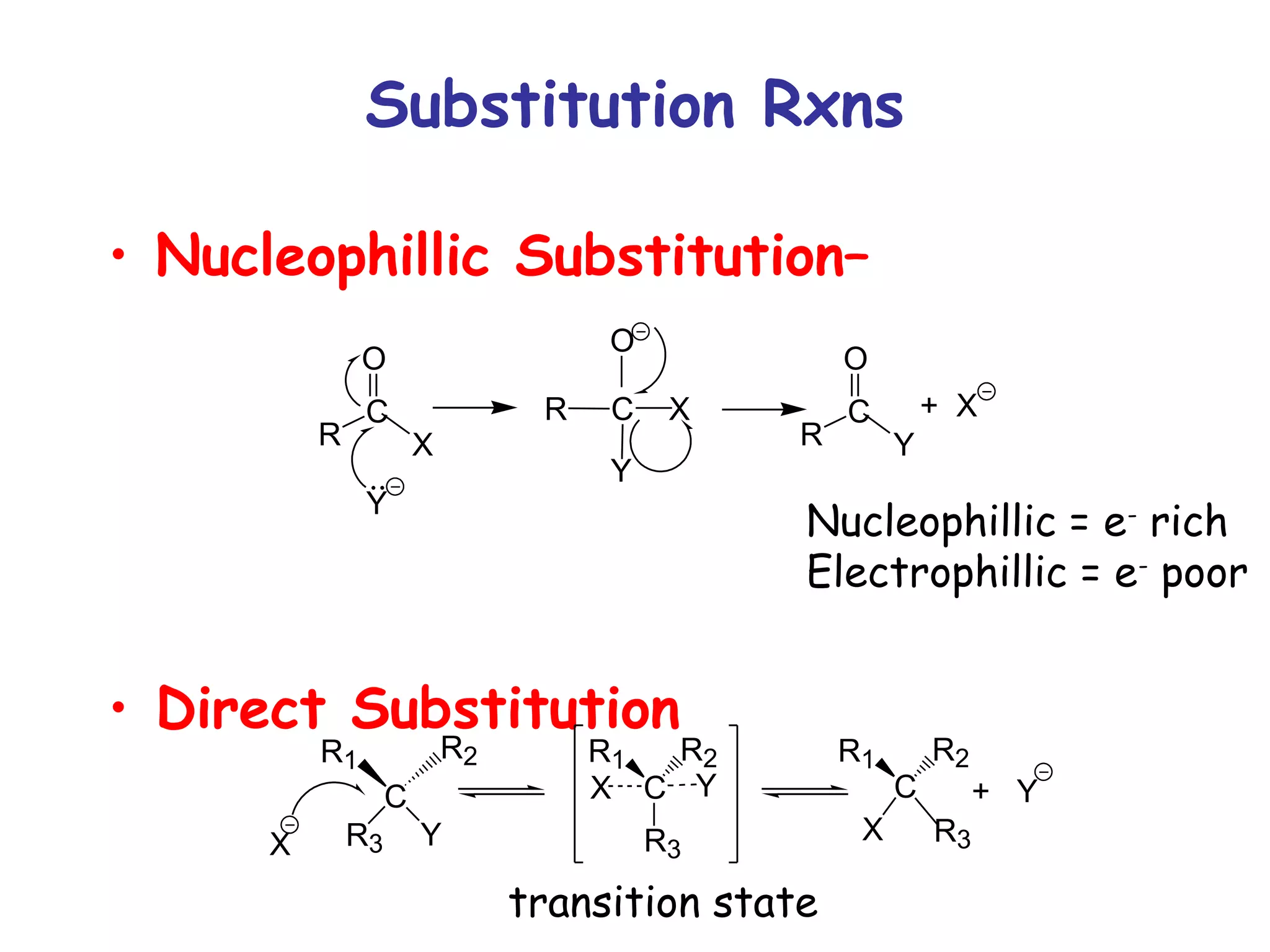
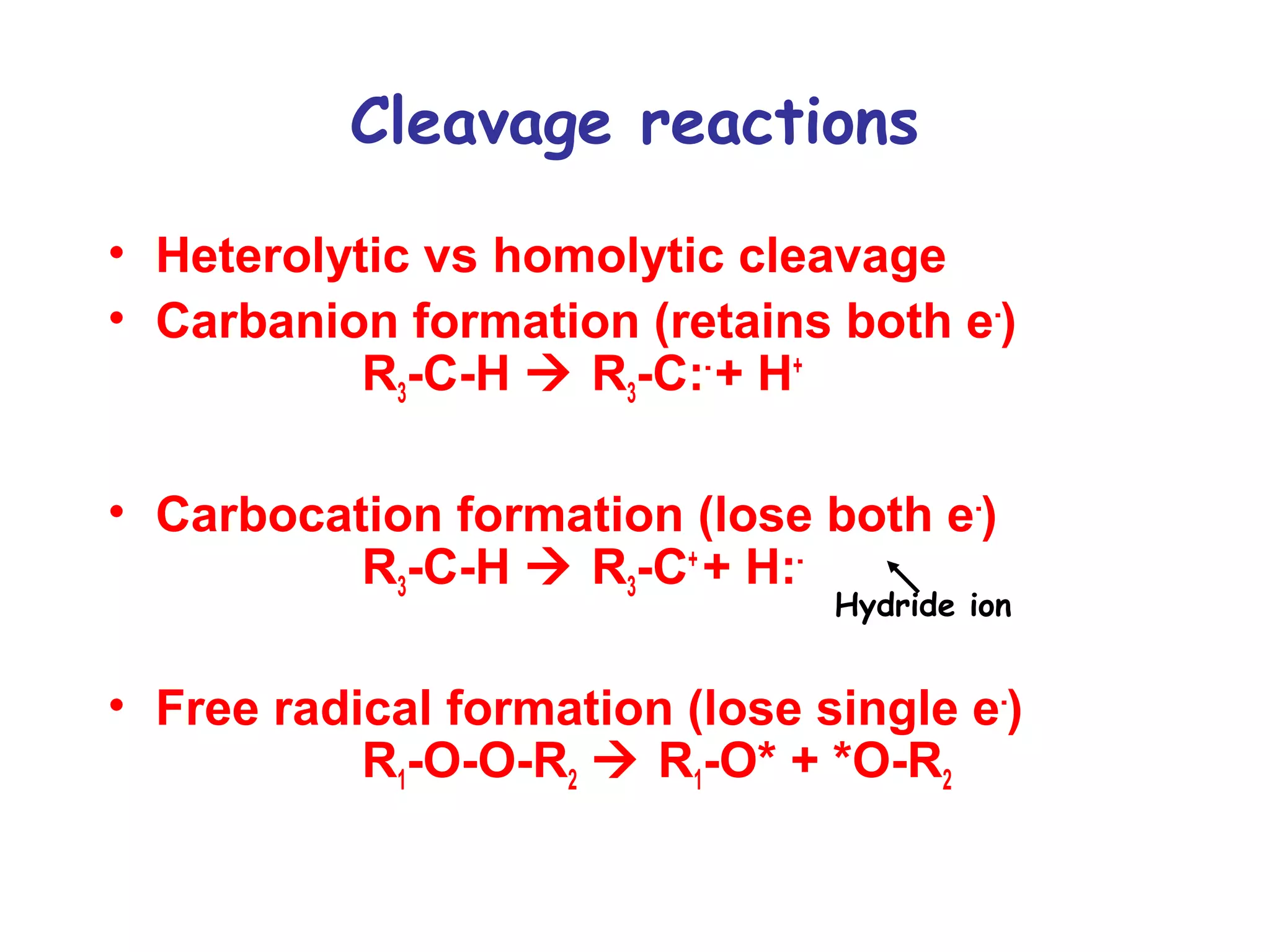
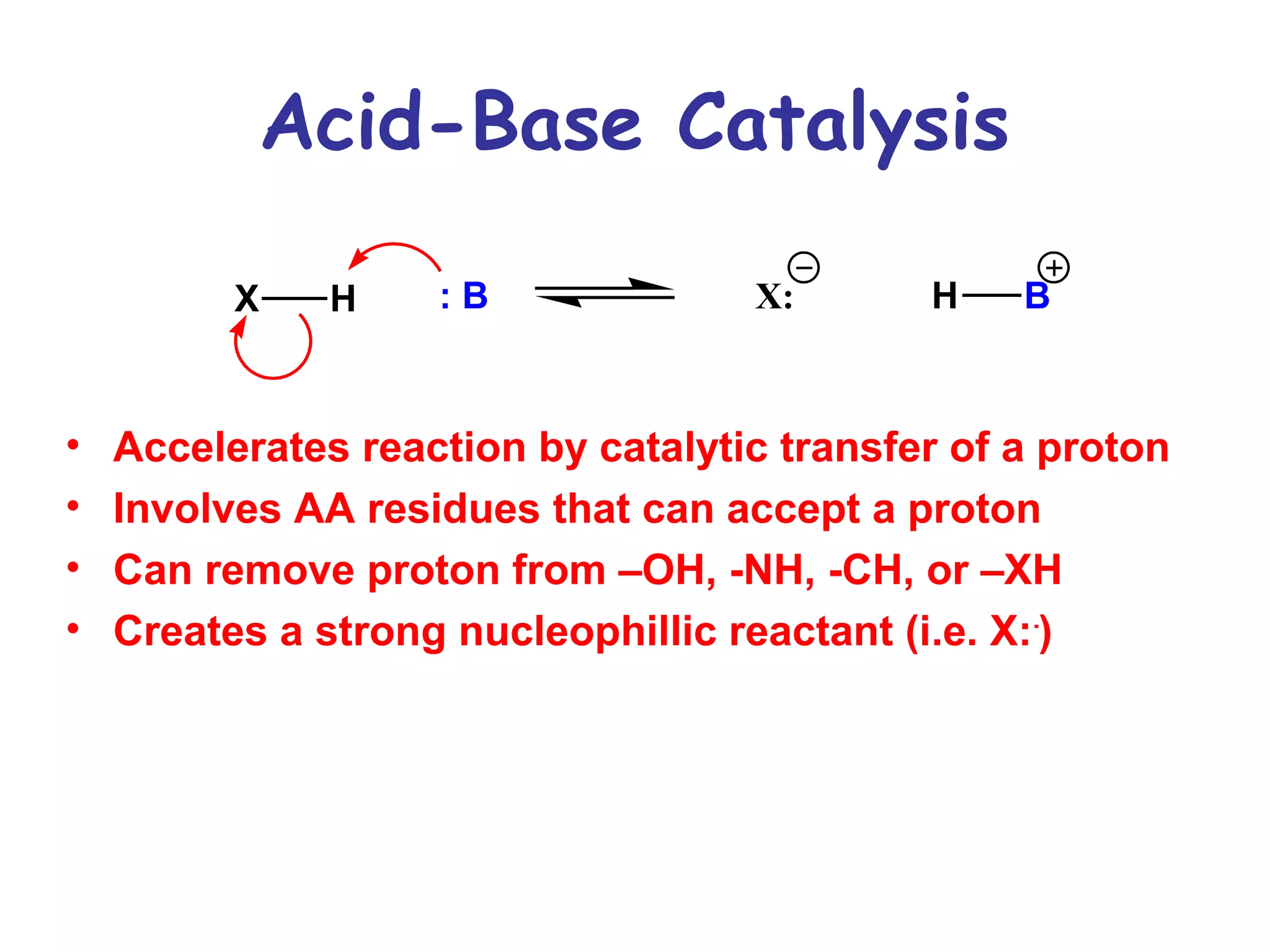
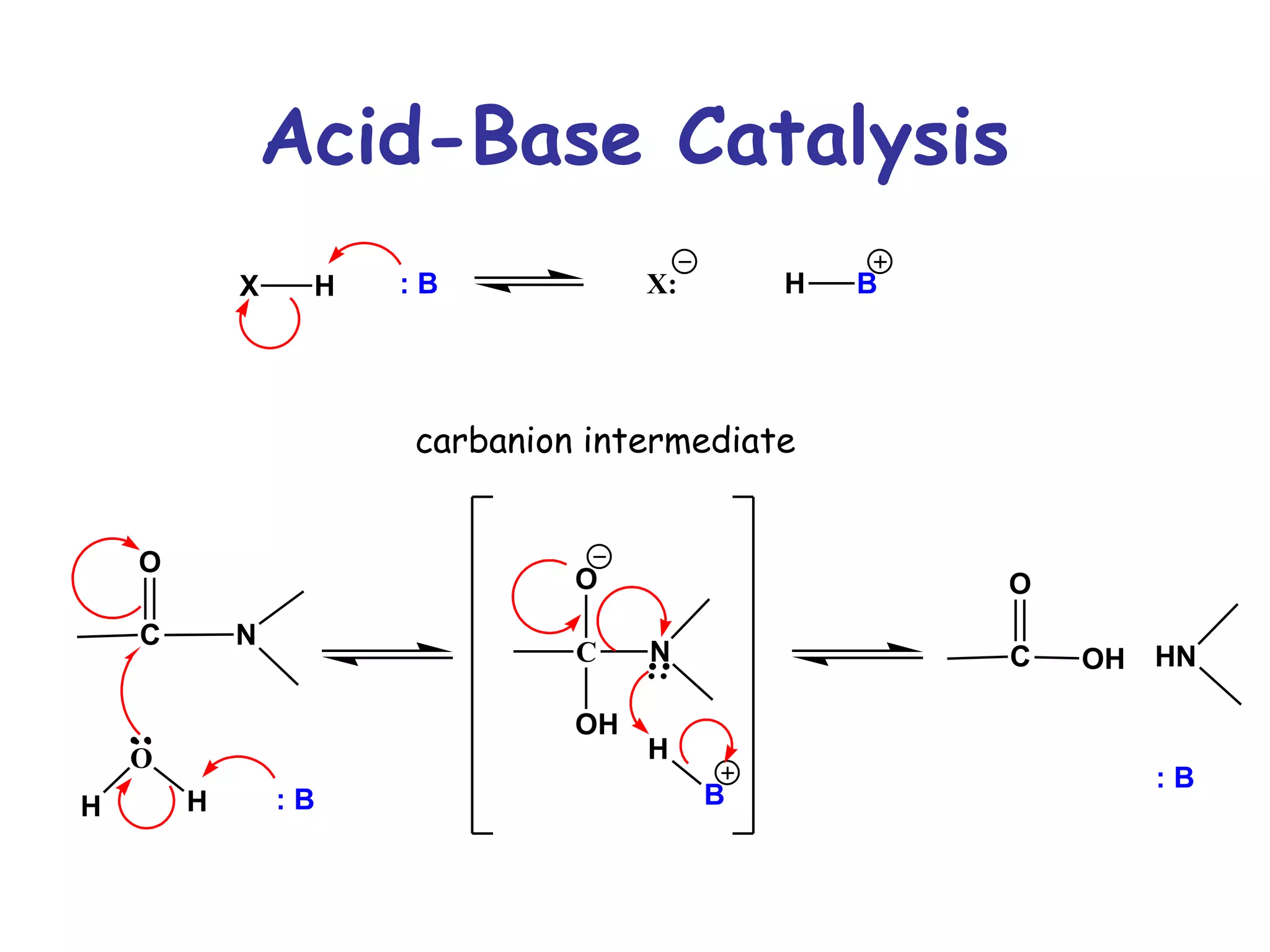
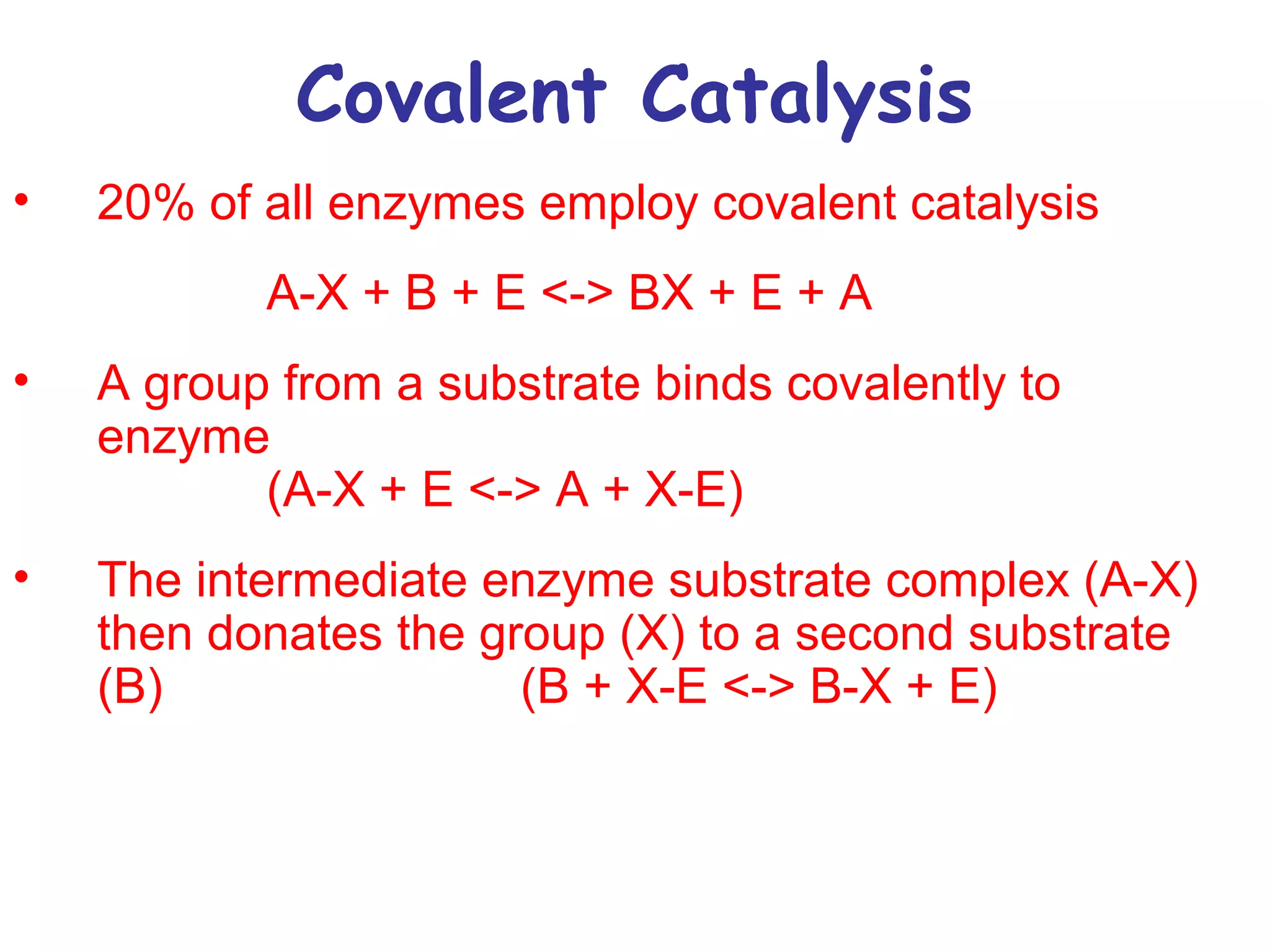
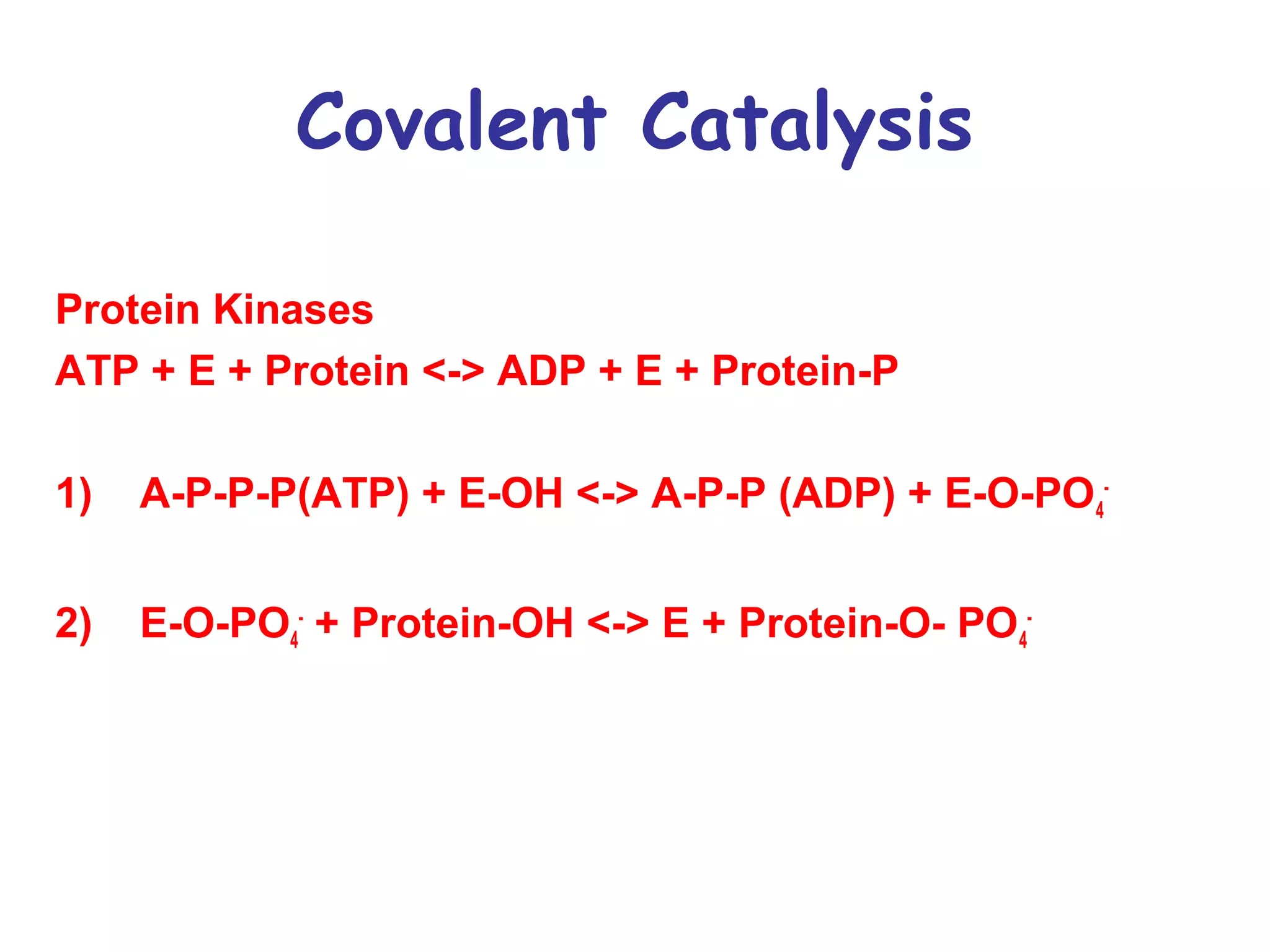
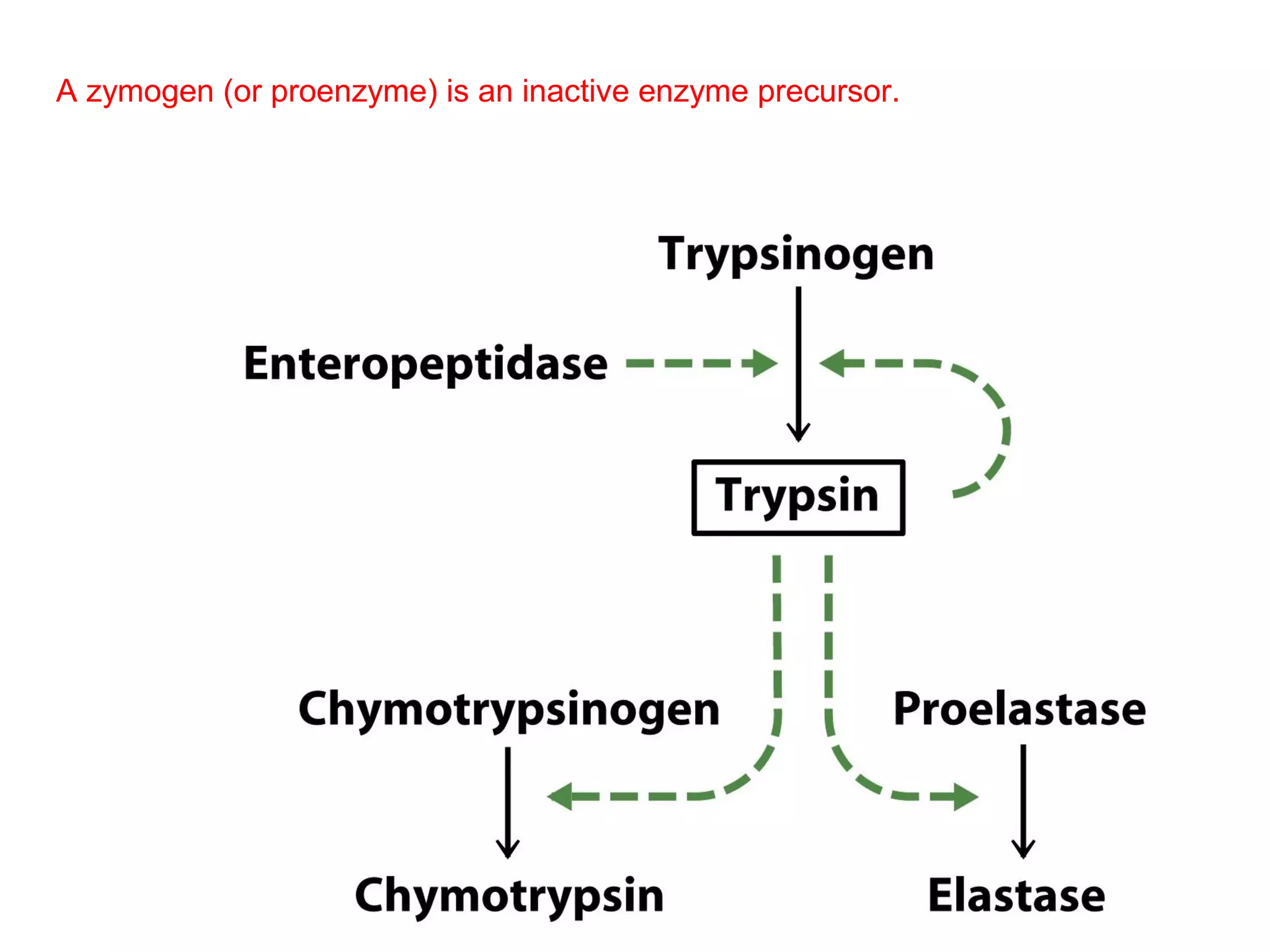
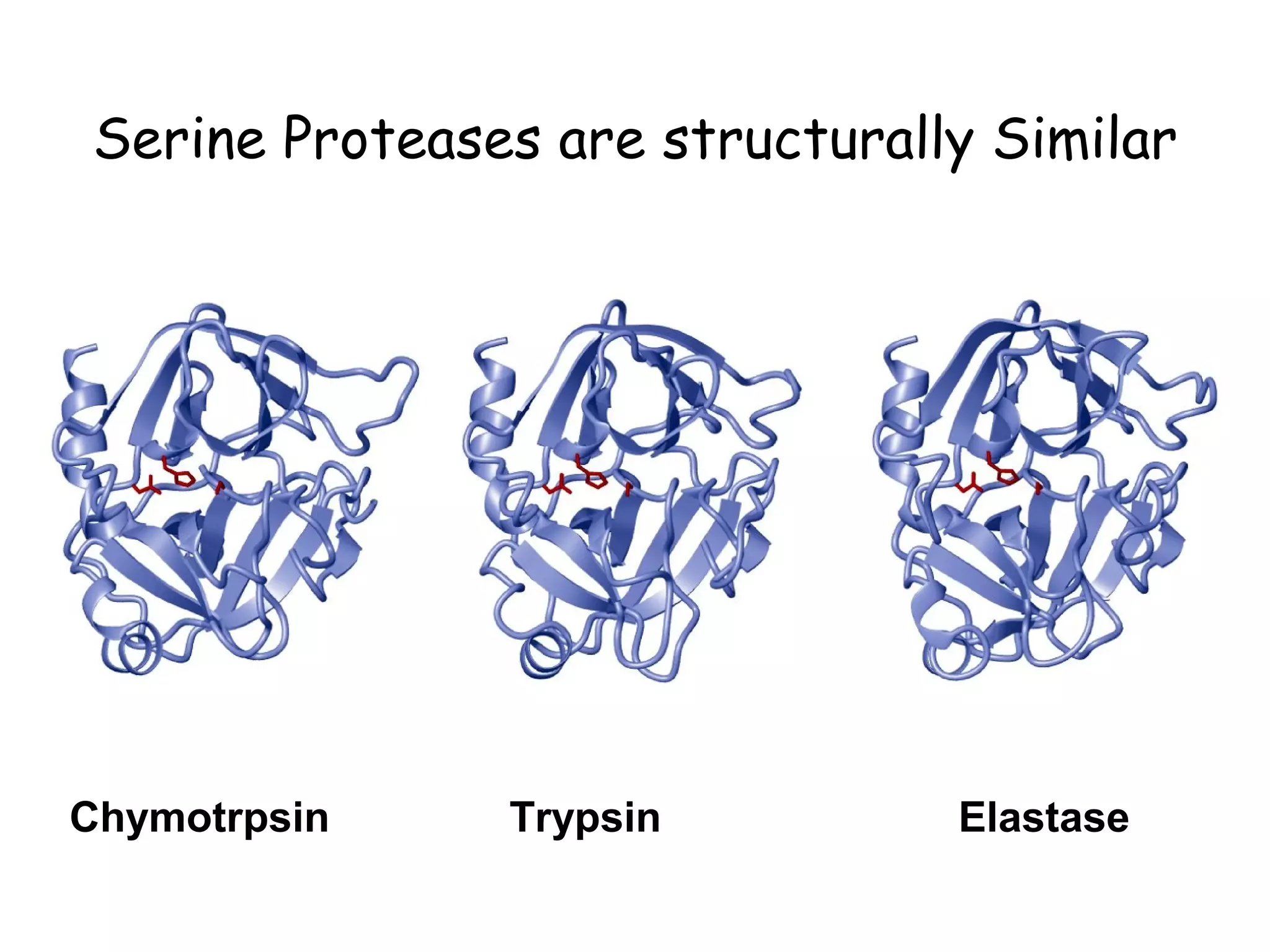
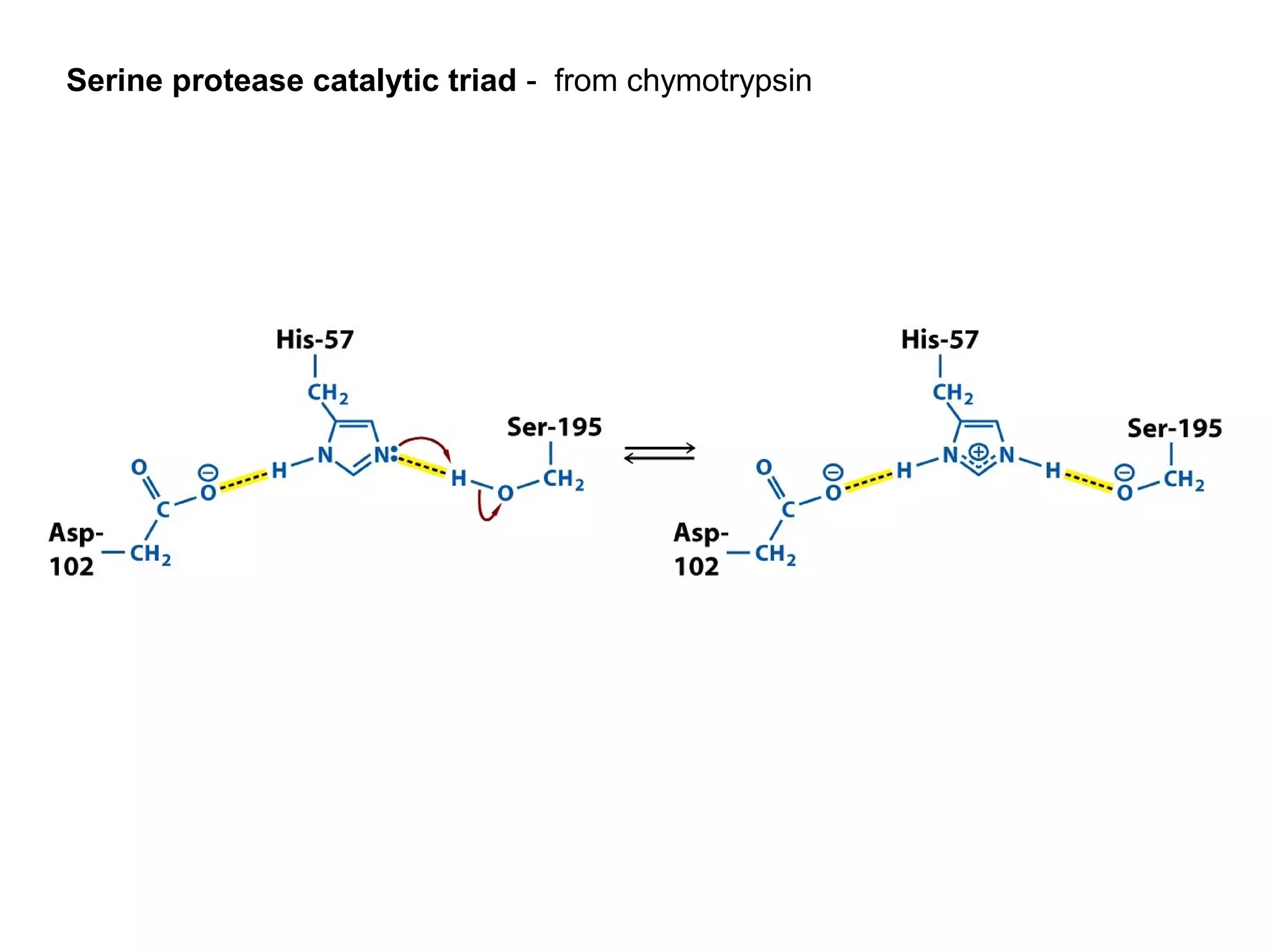
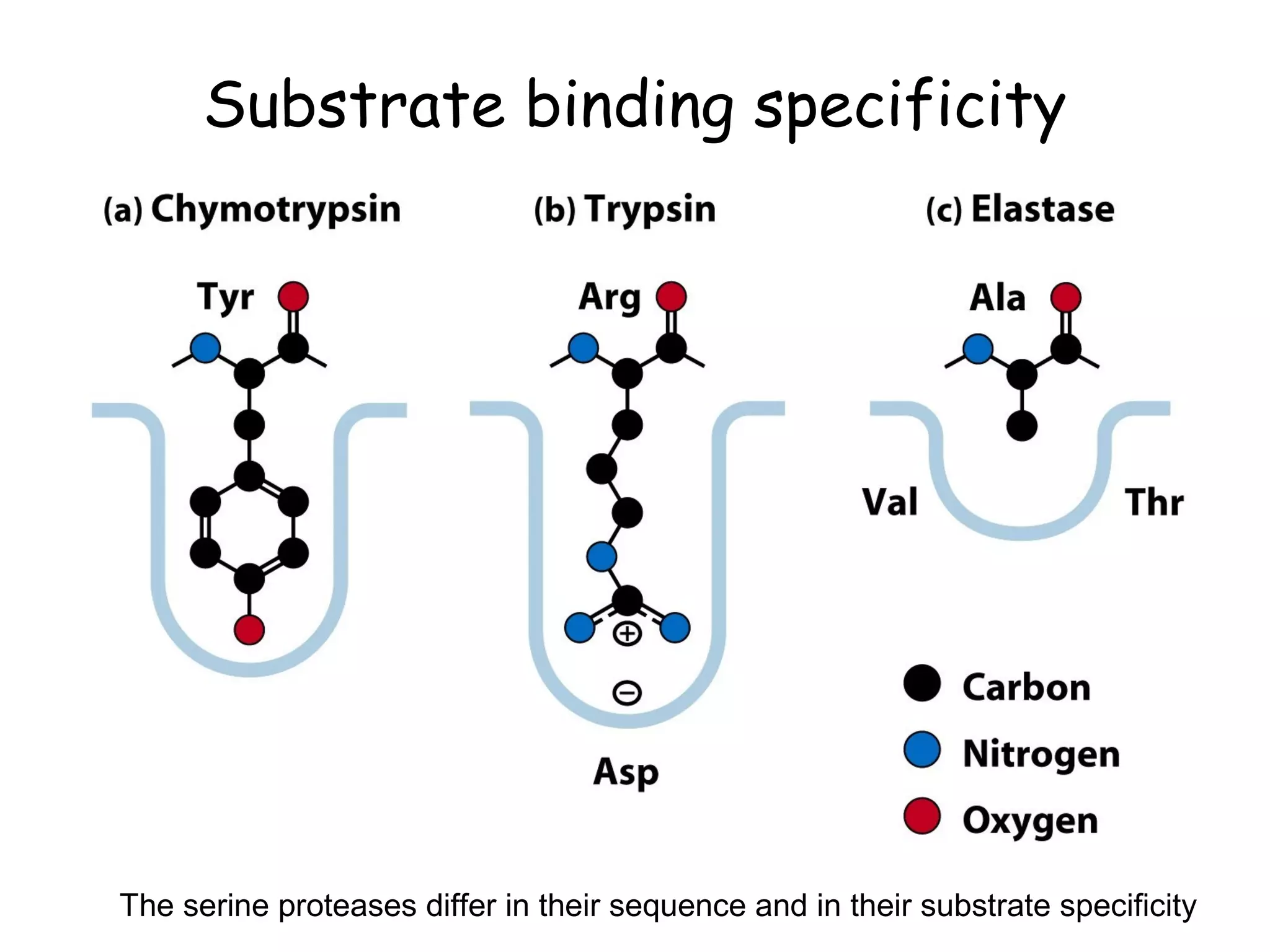
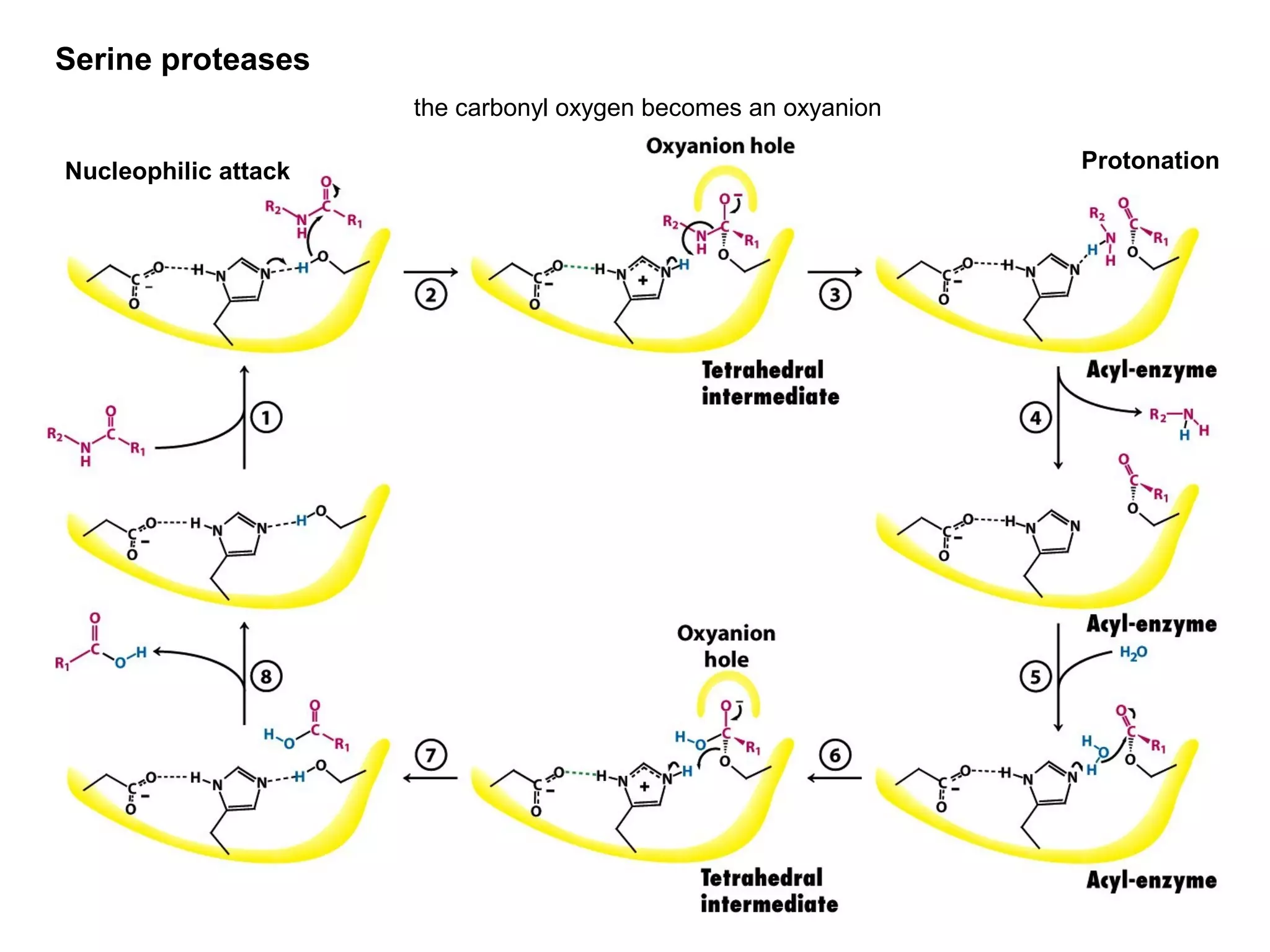
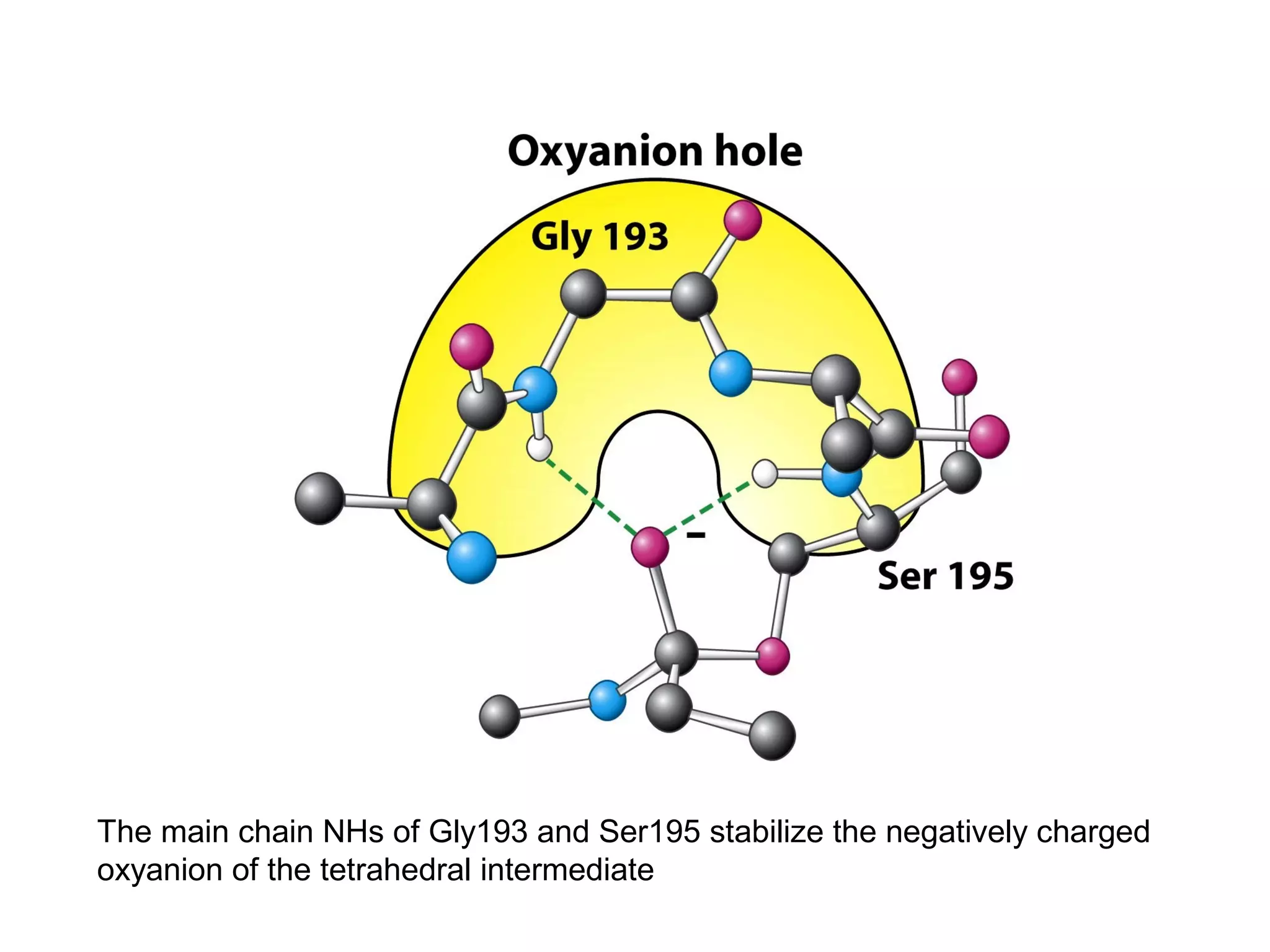
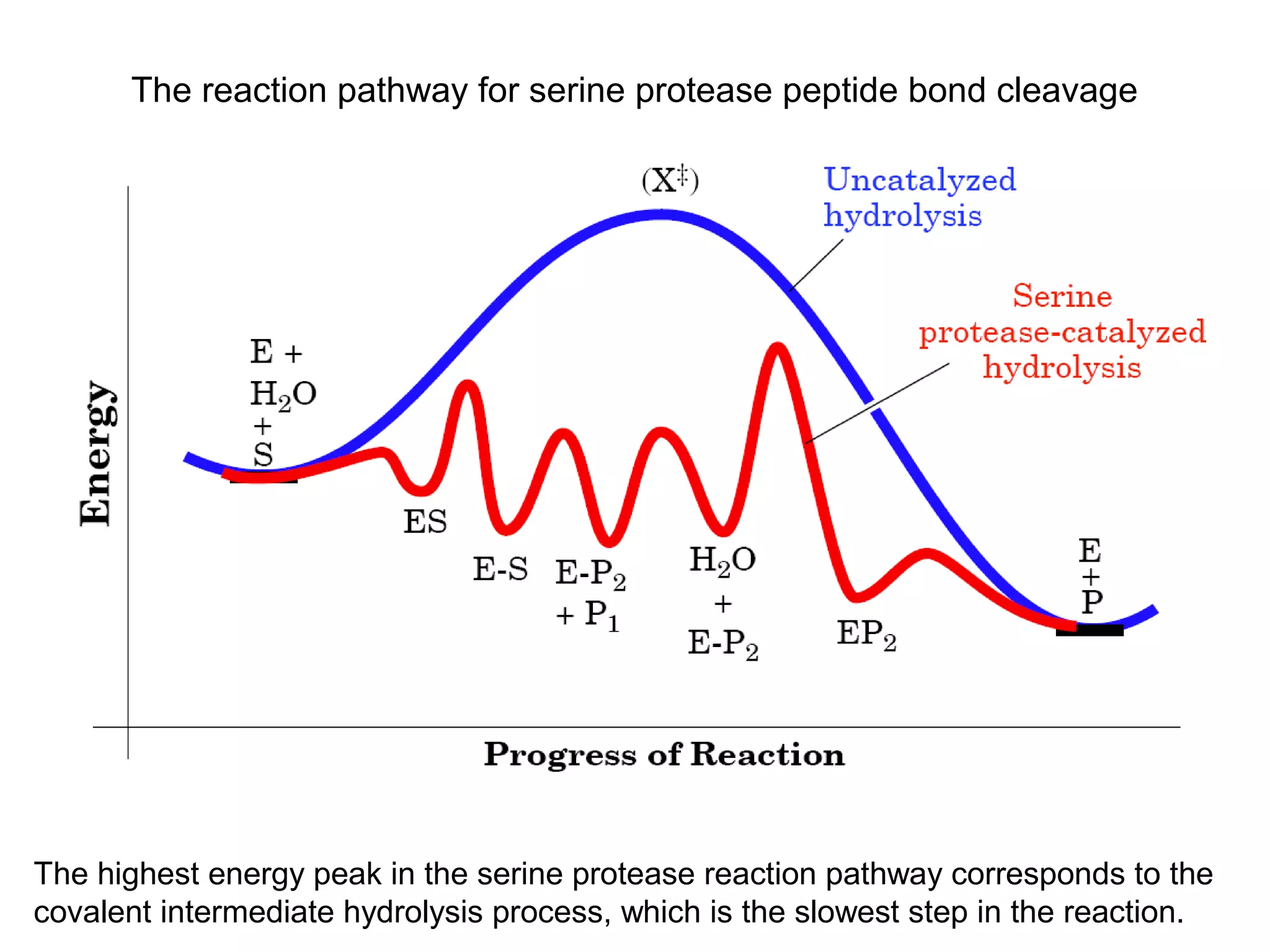
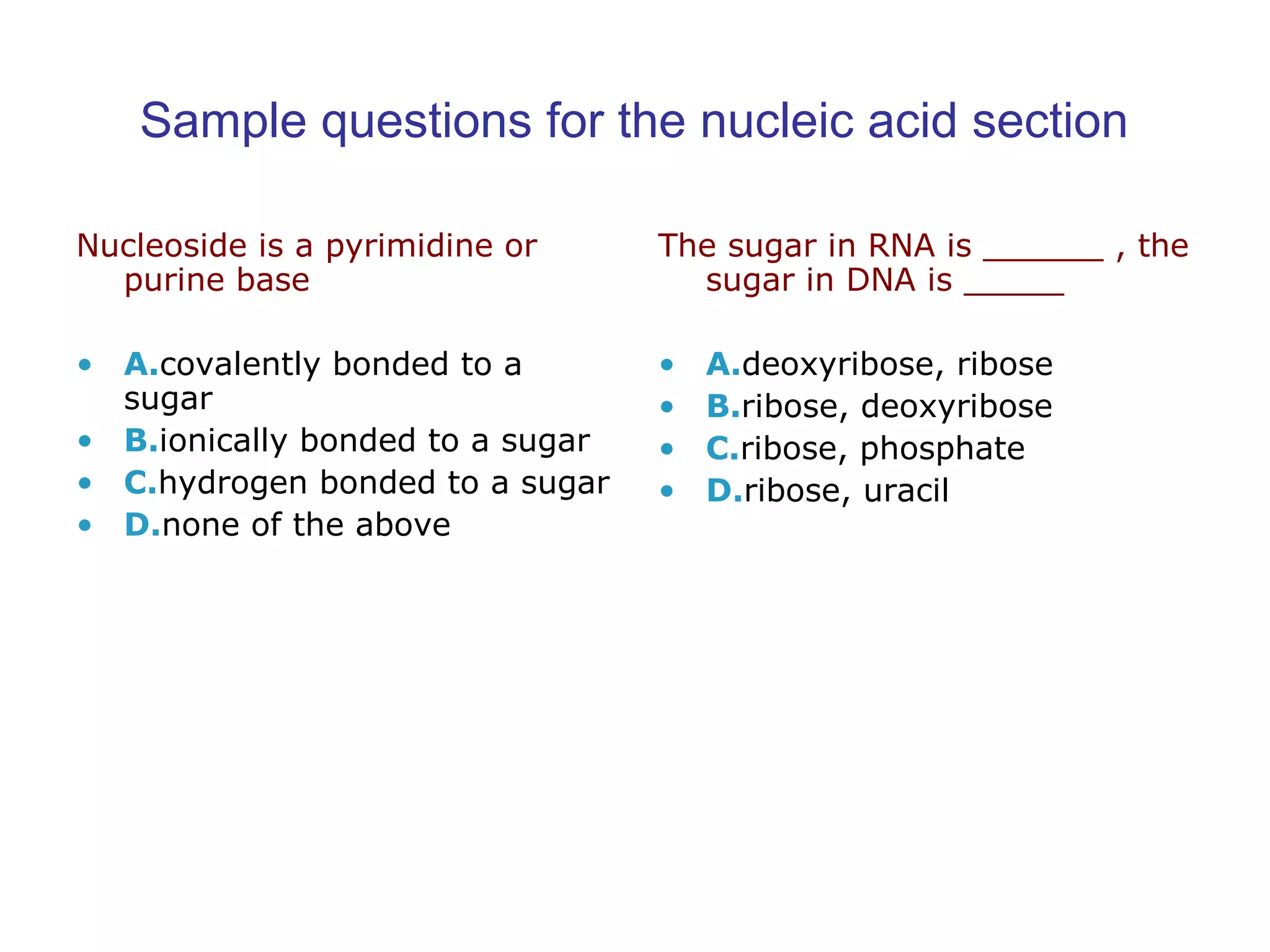
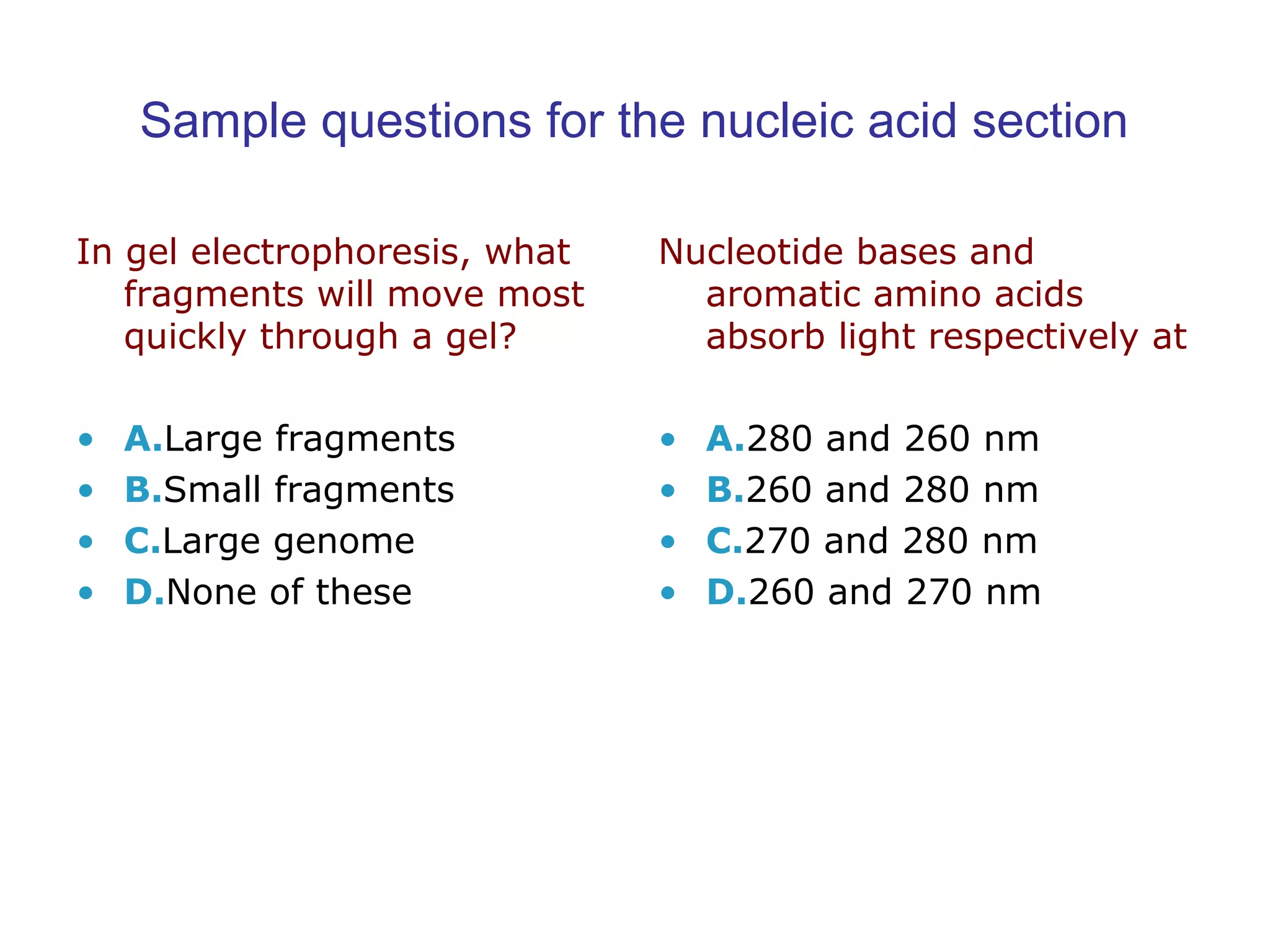
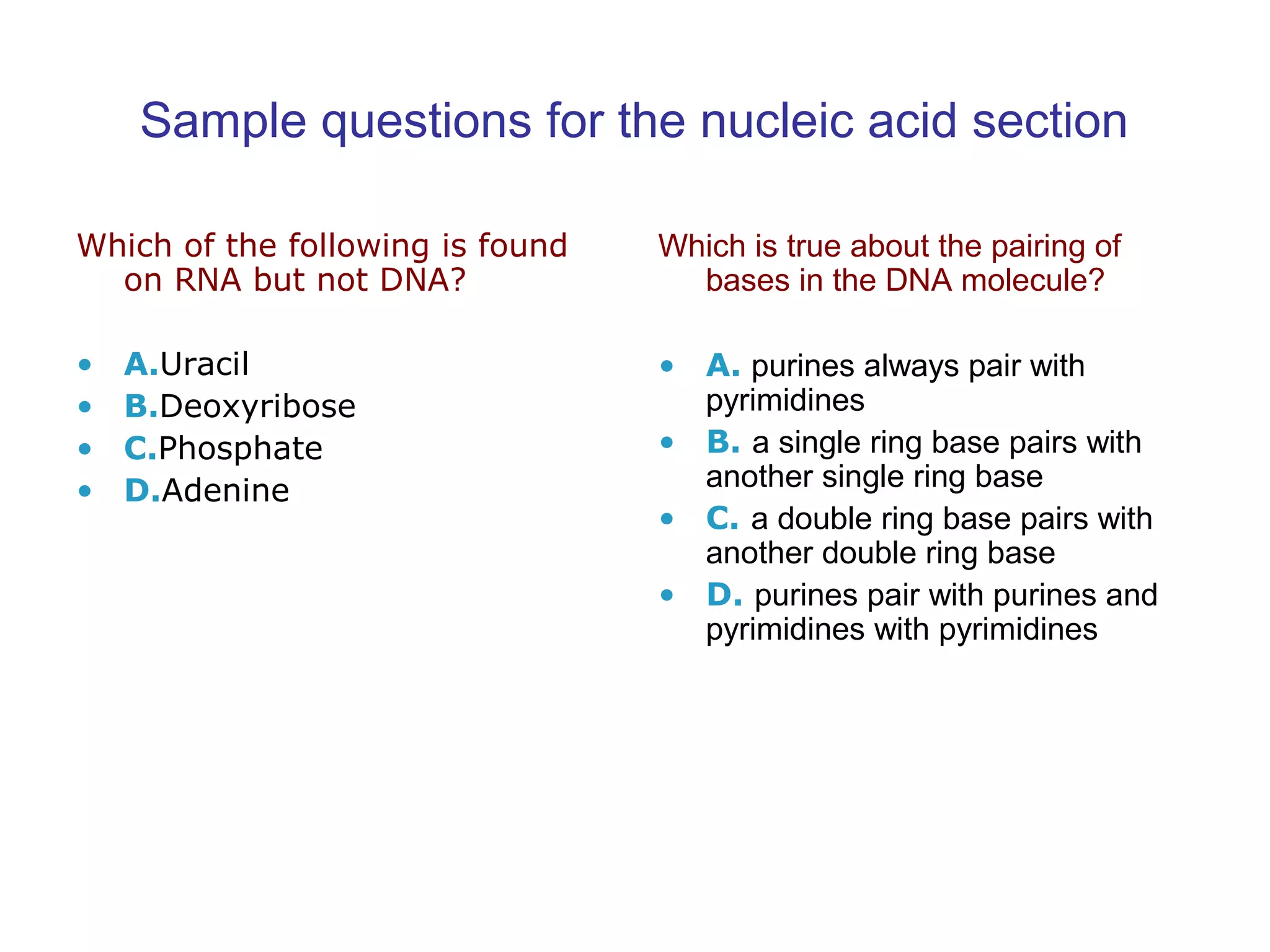
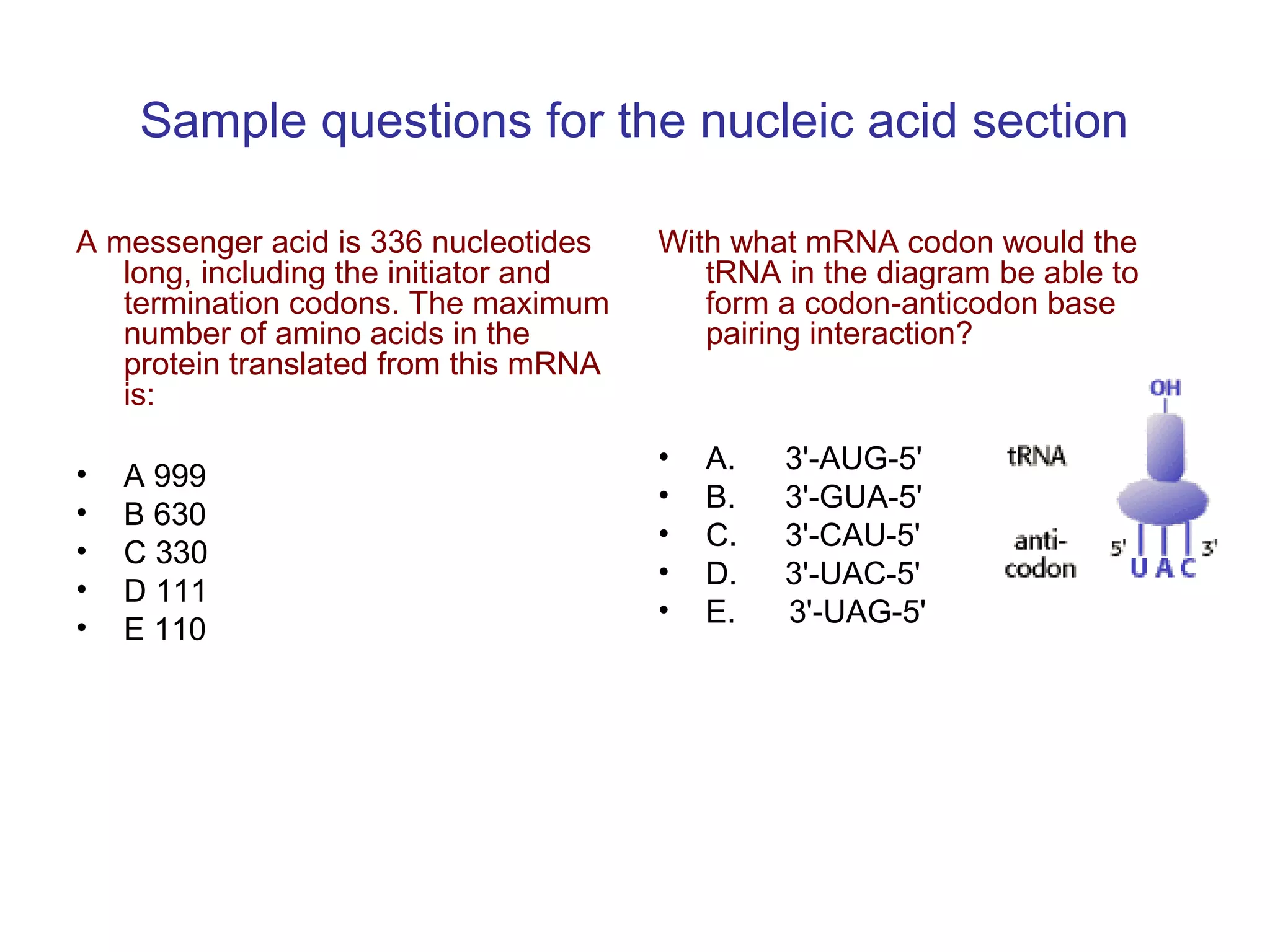
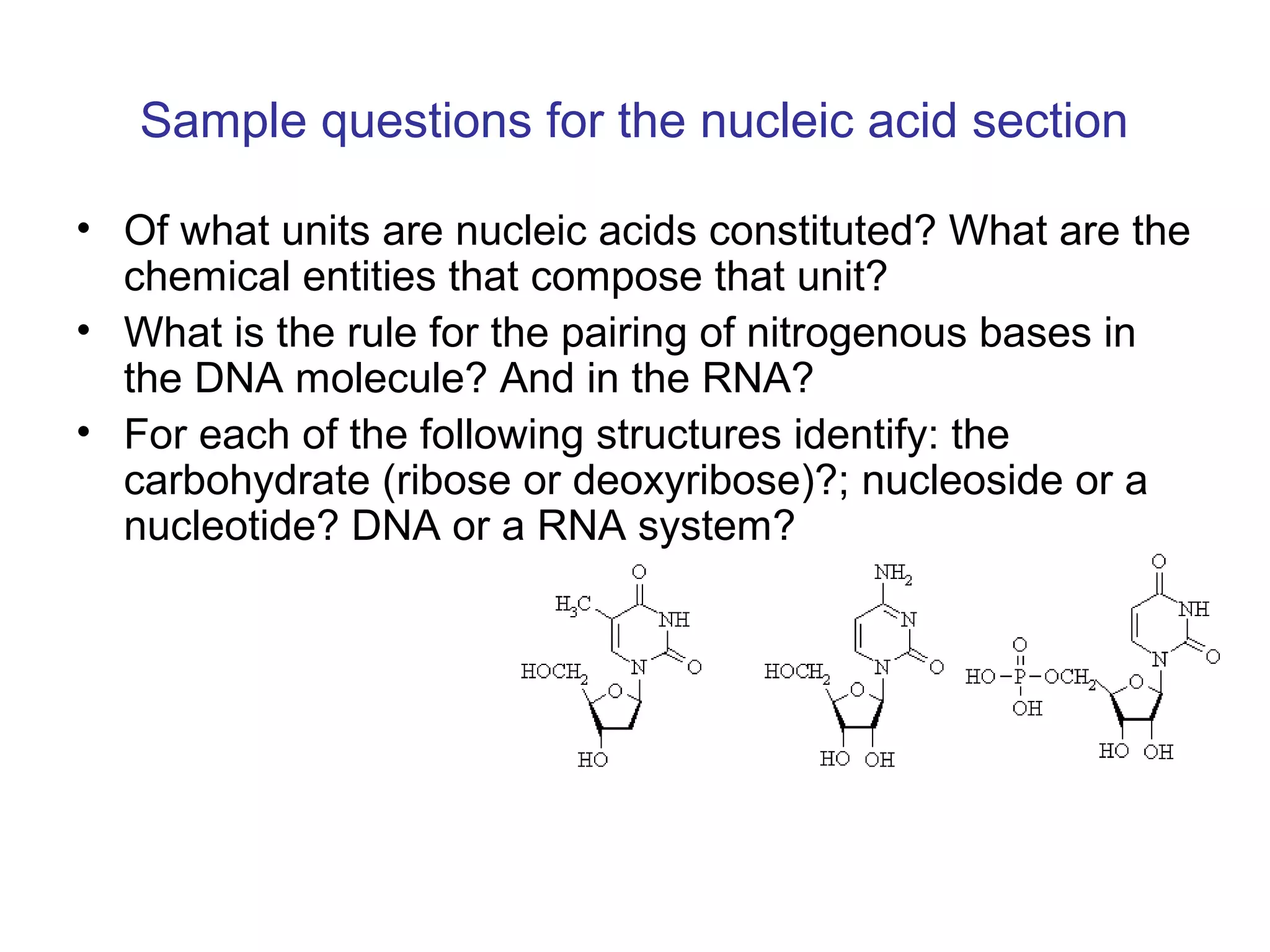
The document contains sample questions about nucleic acids and enzymes. For nucleic acids, it asks about nucleosides, the difference between RNA and DNA sugars, gel electrophoresis of DNA fragments, light absorption of nucleotides and proteins, components found in RNA but not DNA, and base pairing rules. It also includes questions about translating mRNA and forming codon-anticodon interactions. For enzymes, it discusses them as catalysts, mechanisms of catalysis including transition states and active sites, classes of enzymes, coenzymes, and factors affecting enzyme activity. Sample multiple choice questions test understanding of enzyme function, naming, components, effects of temperature, and catalytic mechanisms.

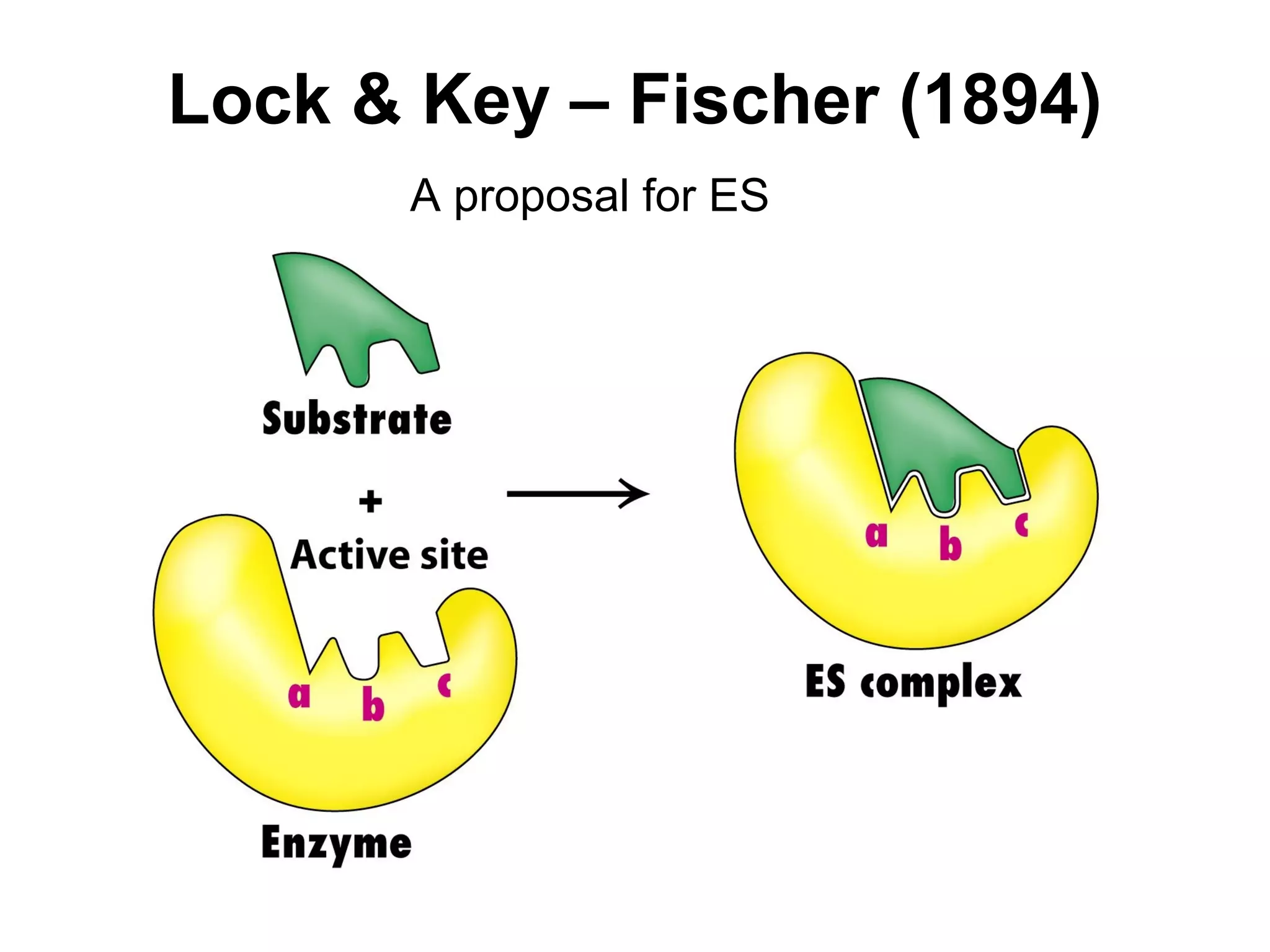
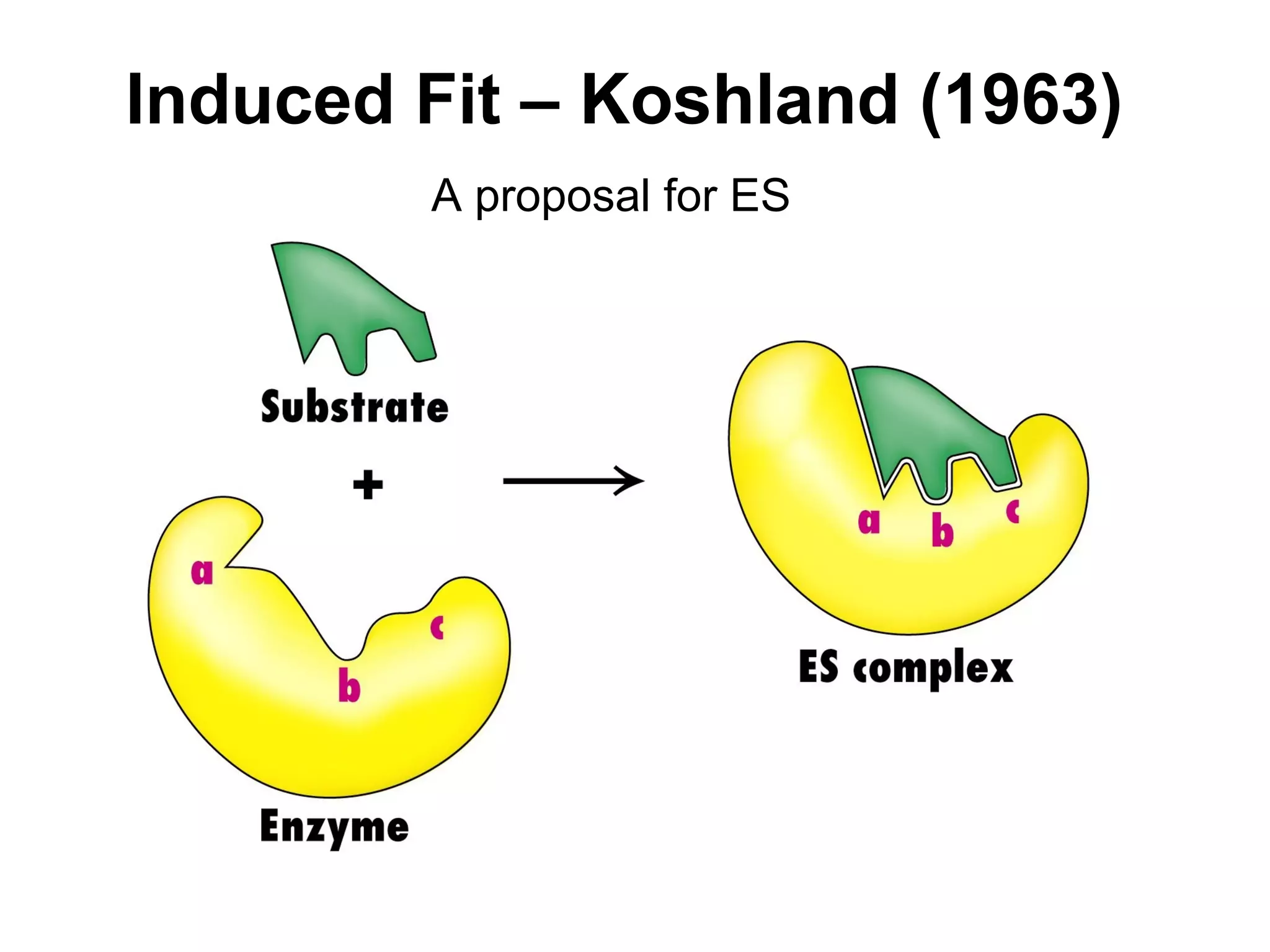
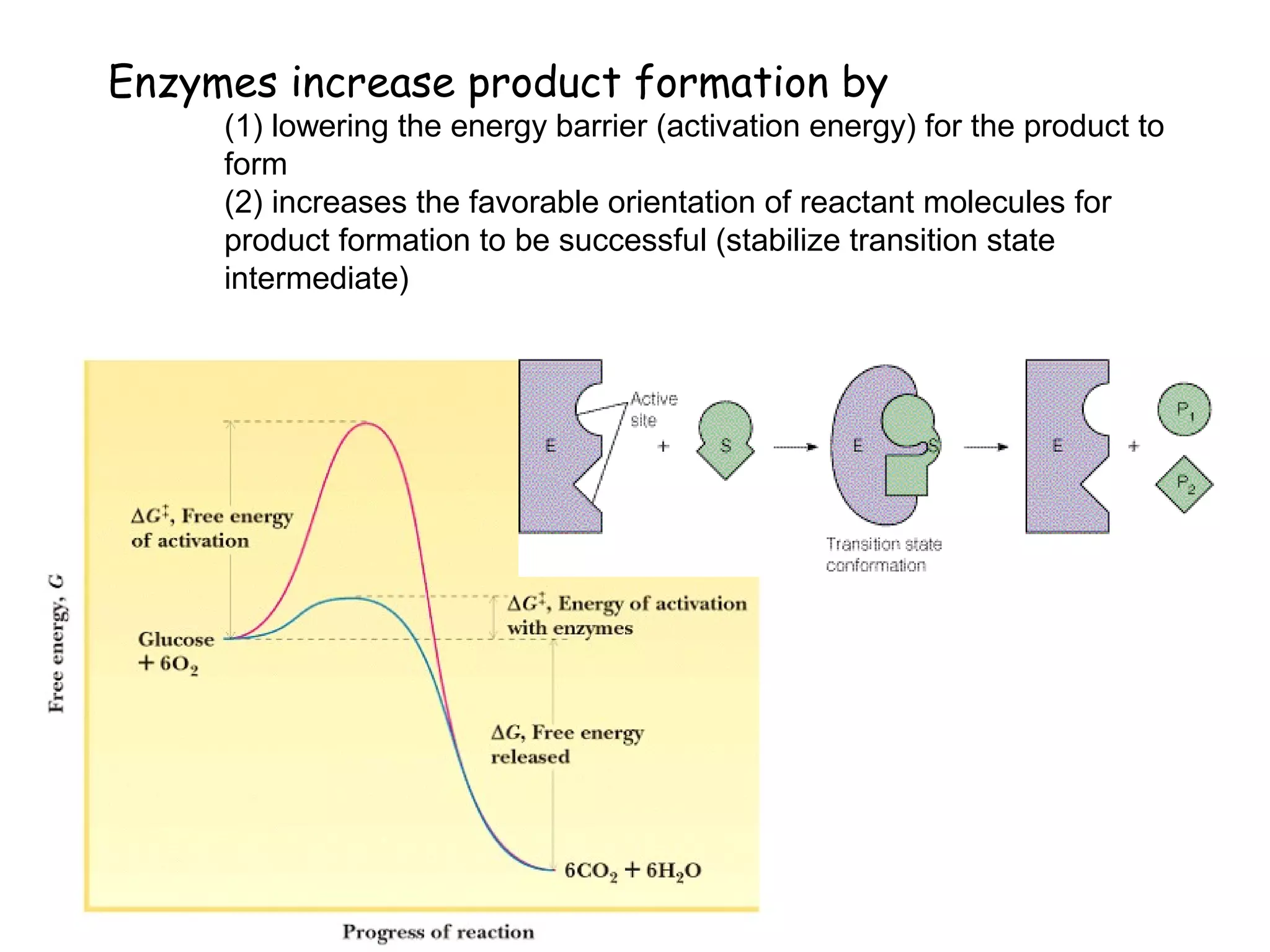
![Enzymatic Catalysis
• Activation Energy (AE) –
The energy require to
reach transition state
from ground state.
• AE barrier must be
exceeded for reaction to
proceed.
• Lower AE barrier, the
more stable the transition
state (TS)
• The higher [TS], the move
likely the reaction will
proceed.
S Ts P](https://image.slidesharecdn.com/wy2m9a0tqson7bdij62q-signature-091447bade647bc70325d98b994f7c3a3901d946a34a4516654ab9d79d1d0b85-poli-141026005446-conversion-gate01/75/Lecture-9-11-2048.jpg)