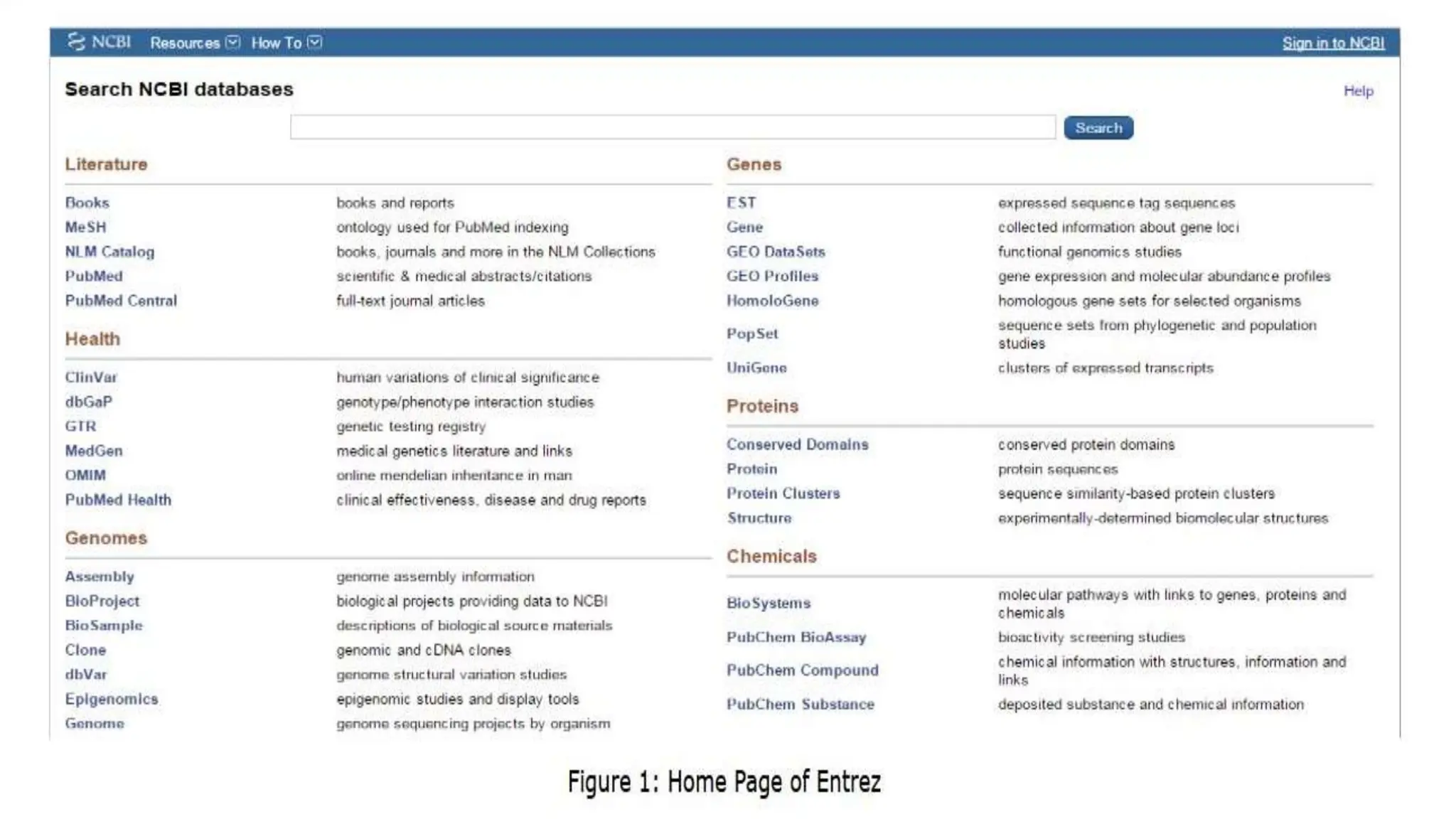
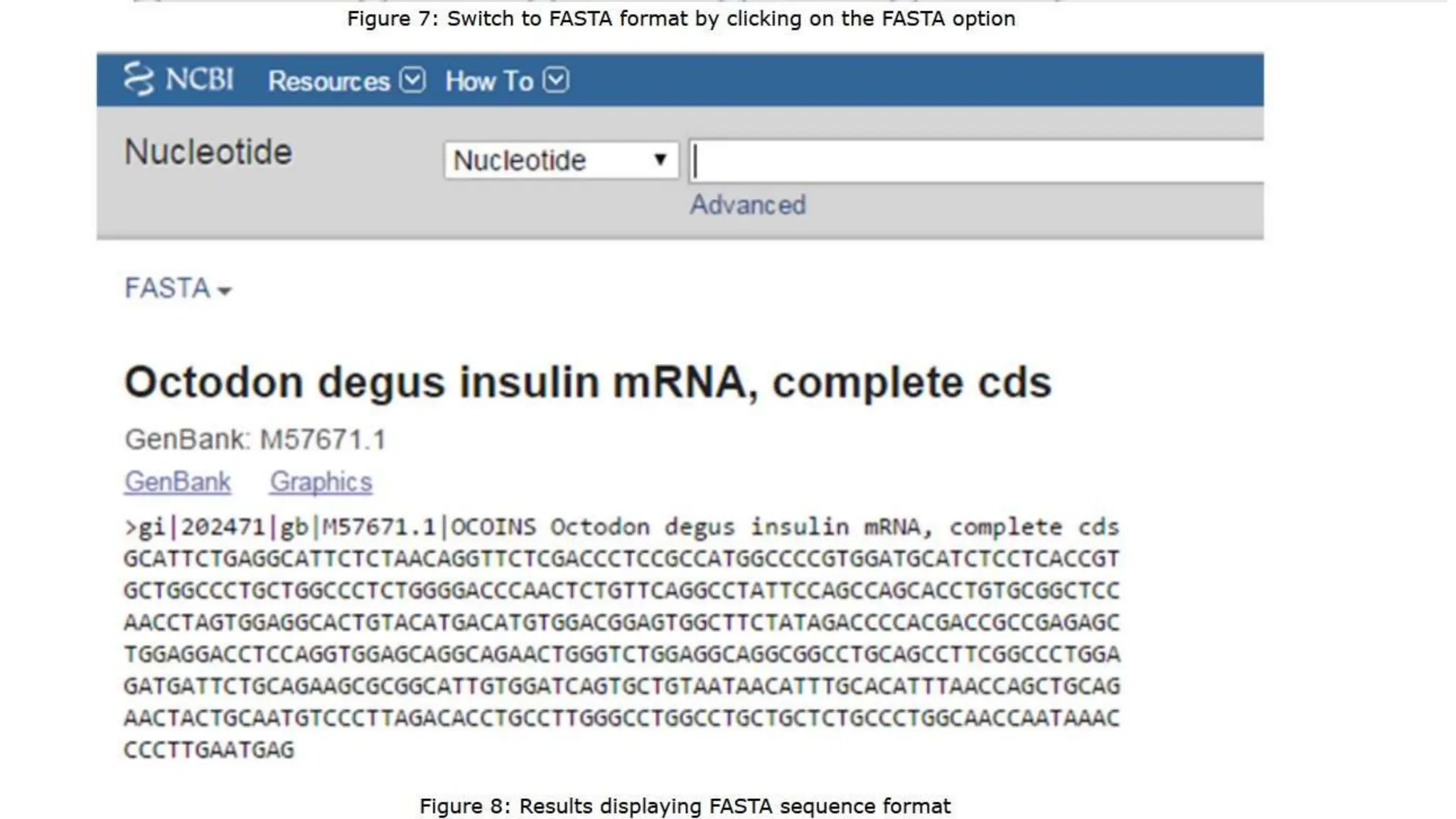
Entrez is a molecular biology database and retrieval system developed by the National Center for Biotechnology Information. It provides an integrated interface to numerous biological databases, allowing users to search across different databases simultaneously. Entrez began as a CD database in 1991 and became available online in 1993. Key features include integrated access to multiple biological databases, batch downloads of large search results, and text-based searching across genetic and bibliographic data using Boolean operators. Users can access Entrez by searching for biological genes, proteins, or other terms.