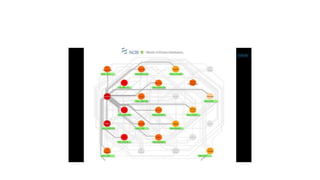
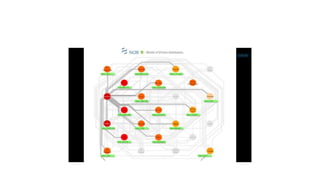
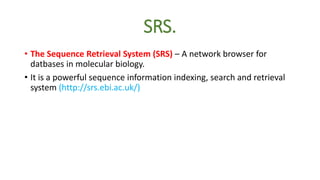
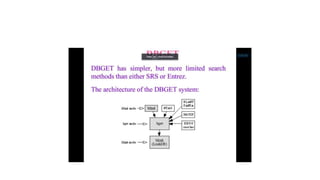
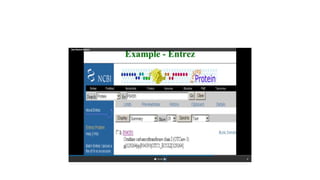
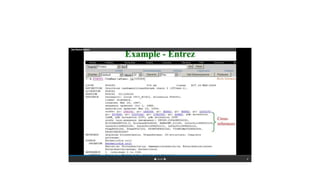
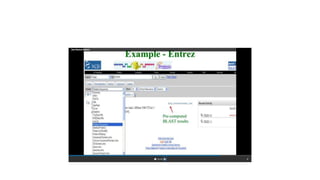
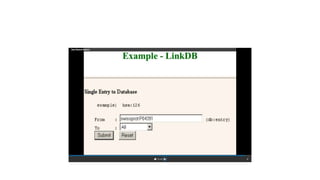
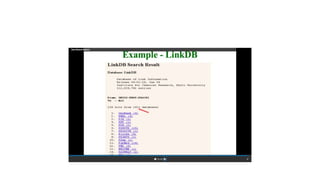
This document describes several text-based biological databases and how to search them. It discusses Entrez, which searches multiple databases and links related entries. It also describes the Sequence Retrieval System (SRS) which allows searching over 80 biological databases. Additionally, it outlines DBGET/LinkDB, an integrated system that searches about 20 databases and links results to associated information. The document provides an example of using each system to retrieve information on a specific protein entry.