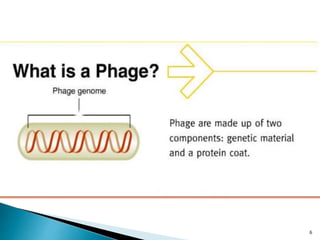
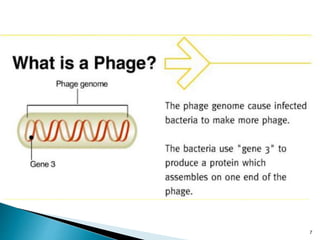
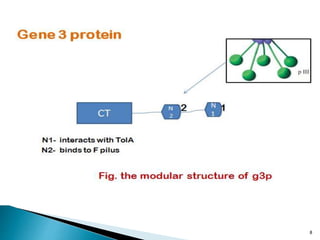
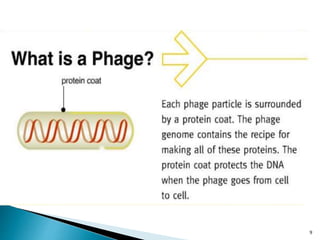
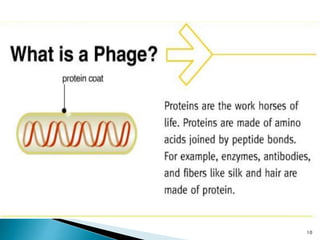
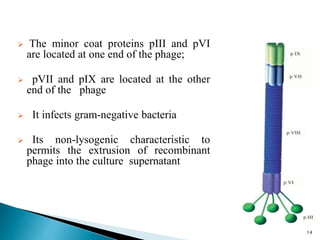
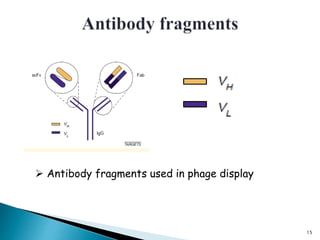
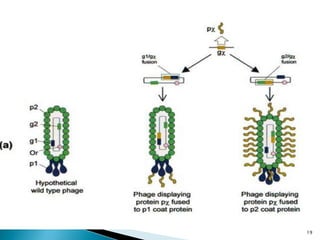
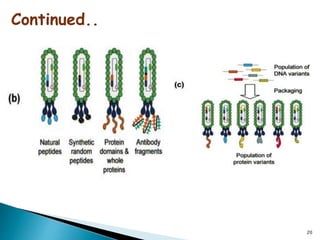
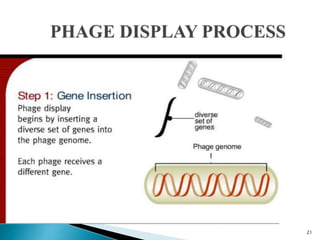
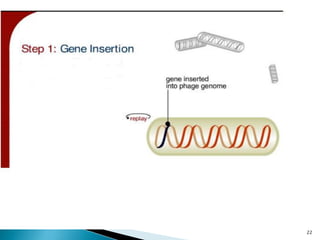
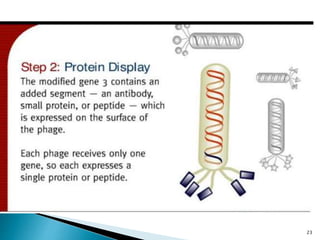
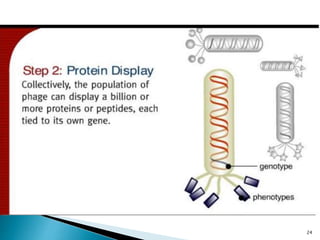
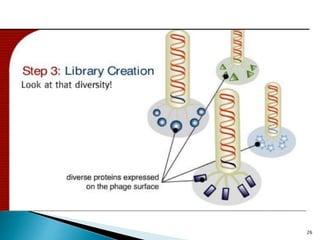
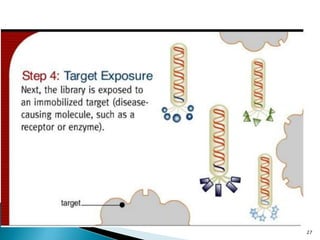
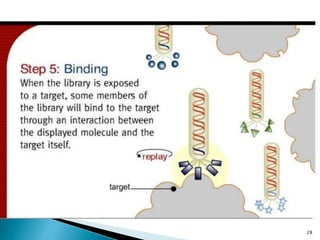
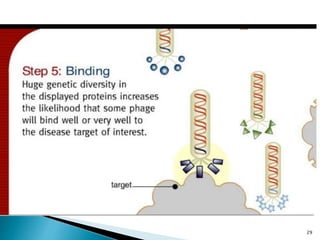
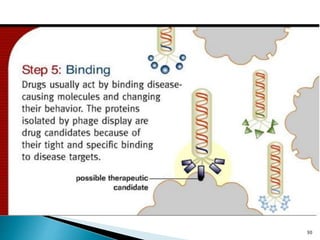
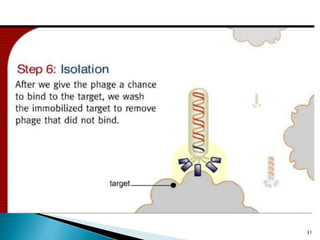
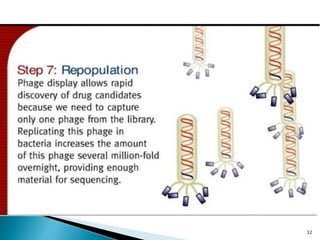
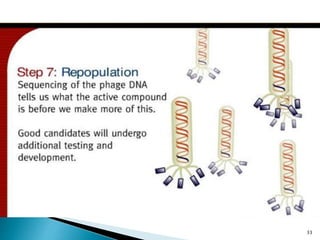
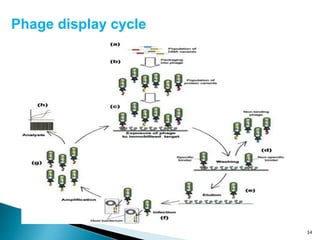
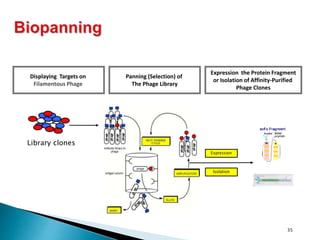
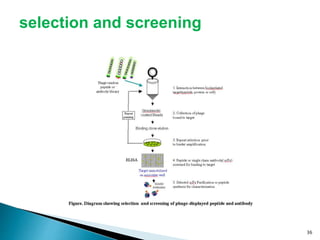
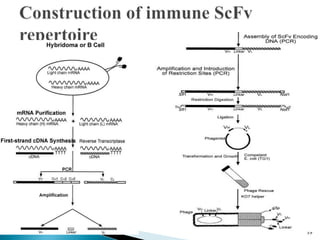
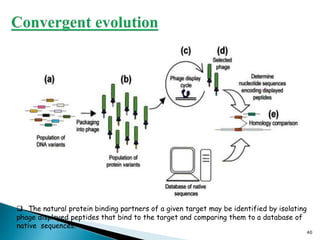
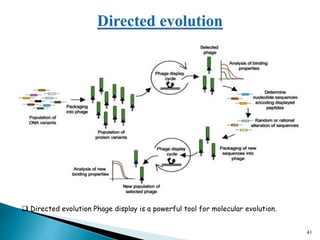
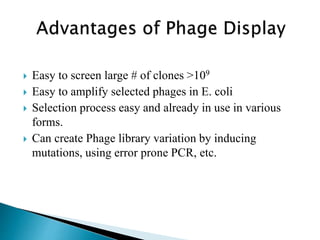
Phage display technology allows the display of proteins or peptides on the outside of bacteriophages while encoding the corresponding gene on the inside. This allows for large libraries to be screened in vitro to select for interactions between the displayed molecules and a target. The most common phages used are filamentous phages like M13, which can be genetically manipulated to display proteins of interest. The technique involves inserting a gene into the phage coat protein gene, infecting bacteria to produce phage particles displaying the protein, and panning against a target to isolate interacting proteins.