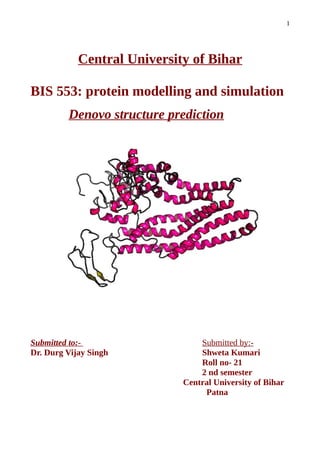
This document discusses de novo protein structure prediction, which predicts protein structure from amino acid sequence alone without using existing protein templates. It notes the need for ab initio prediction when no homologous structures exist. Successful de novo prediction requires an accurate energy function to identify native structures, an efficient conformational search method, and ability to select native models. Results from ab initio prediction typically have 5-10 Angstrom accuracy. Domain prediction is important to divide large proteins into independently folding domains for prediction. Advantages include automation and ability to structurally annotate genomes. Challenges include the vast conformational search space and need for accurate energy functions.