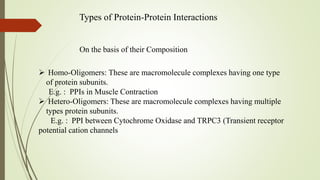
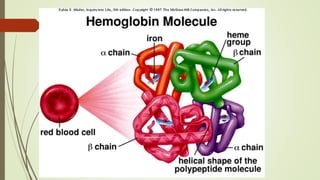
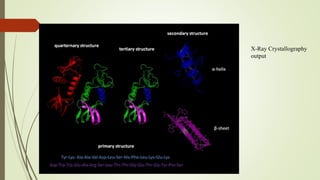
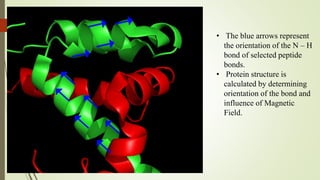
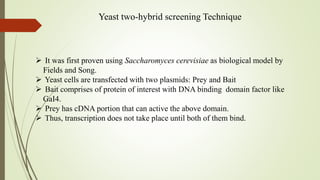
The document discusses protein-protein interactions (PPIs), including an introduction to PPIs, the types of interactions, techniques used to study them like X-ray crystallography, NMR spectroscopy and cryo-electron microscopy, and factors that affect PPIs. It also covers methods to investigate PPIs such as affinity purification coupled with mass spectrometry and yeast two-hybrid screening. Applications of understanding PPIs include developing therapeutic drugs and identifying functions of unknown proteins.