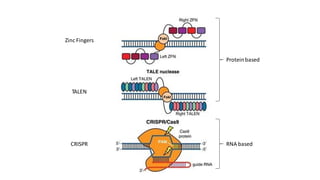
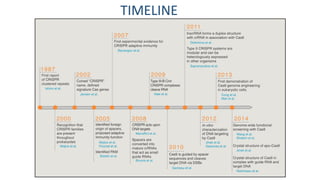
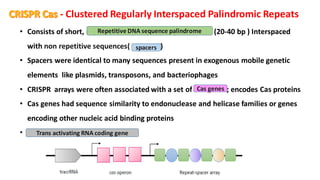
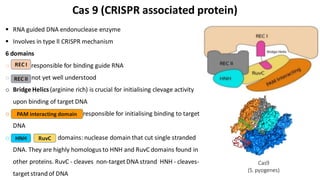
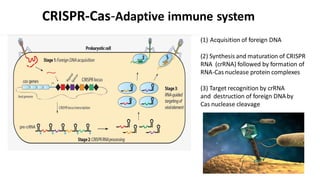
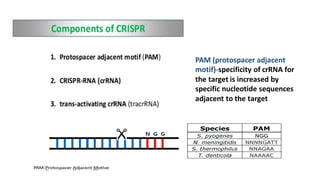
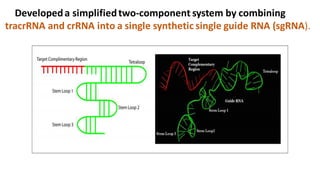
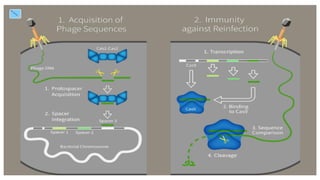
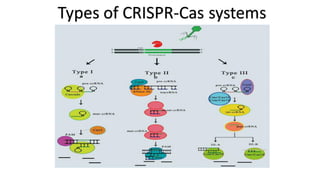
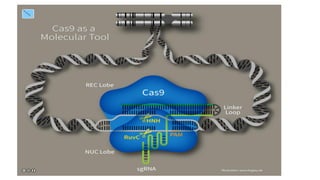
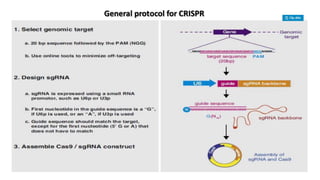
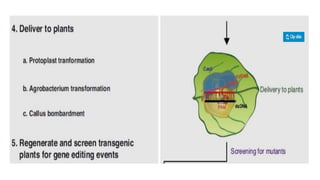
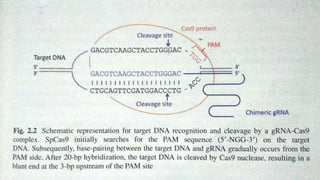
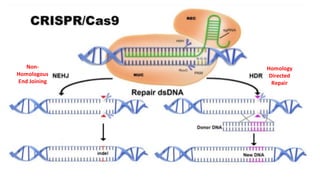
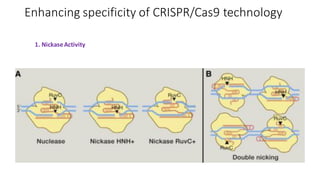
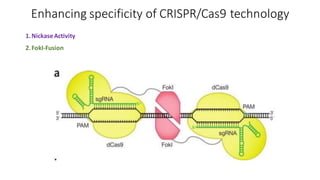
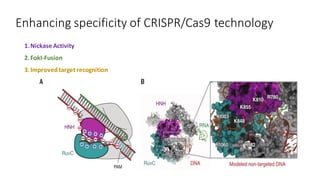
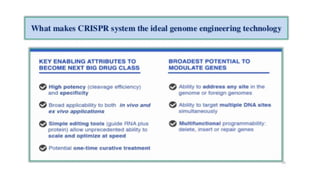
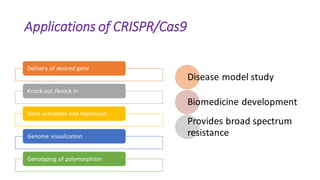
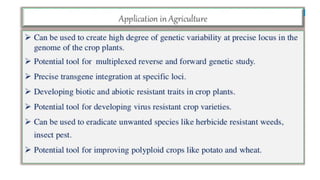
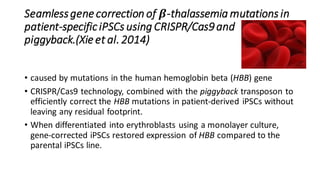
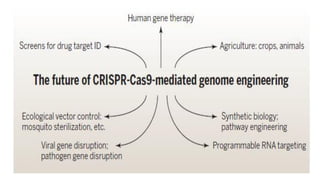
CRISPR/Cas9 is a powerful genome editing tool that allows genetic material to be added, altered or removed at specific locations in the genome. It involves a bacterial adaptive immune system where CRISPR sequences and Cas genes work together. The Cas9 protein uses a guide RNA to introduce double stranded breaks at targeted DNA sequences. This enables precise genome editing through non-homologous end joining or homology directed repair. CRISPR/Cas9 provides a simple and accurate way to modify genes for applications in research, medicine, agriculture and more. While it holds great promise, there are also limitations and concerns regarding off-target effects that researchers continue working to address.