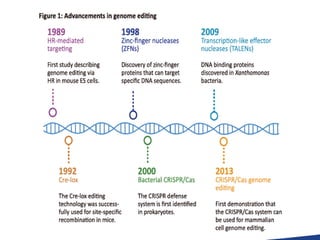
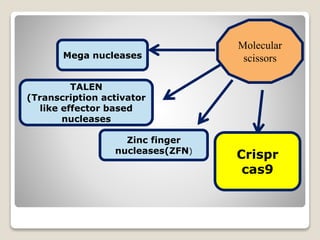
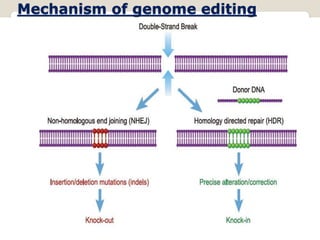
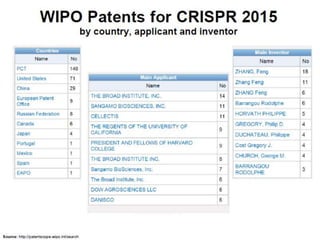
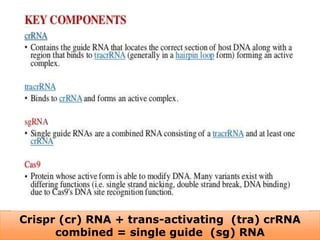
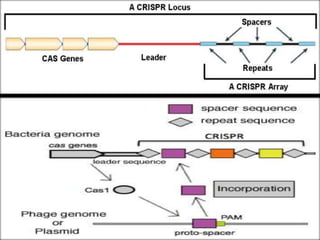
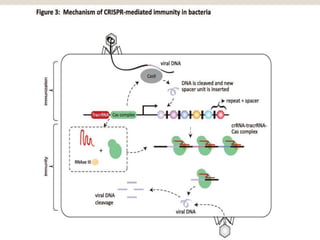
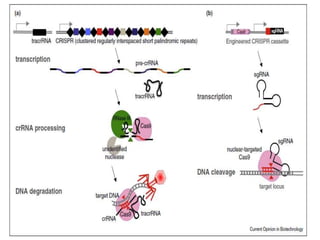
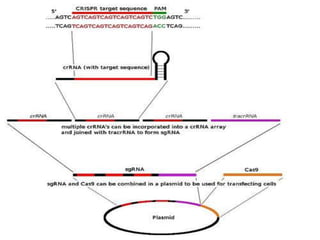
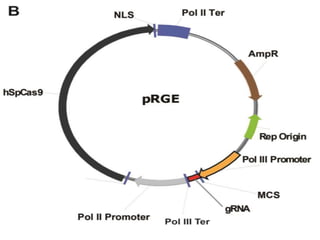
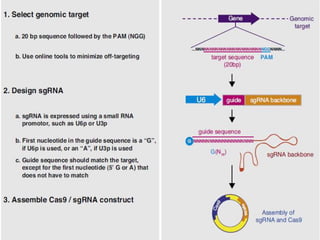
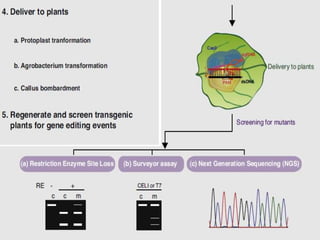
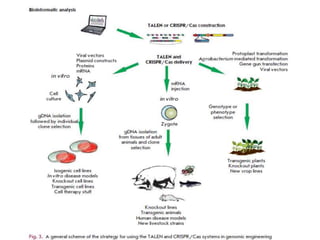
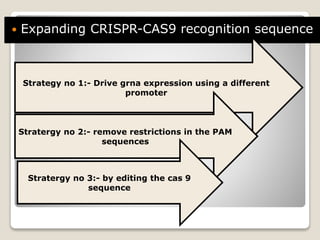
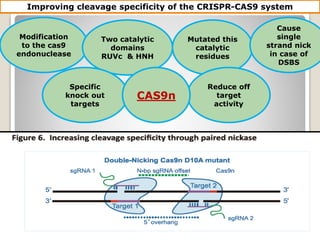
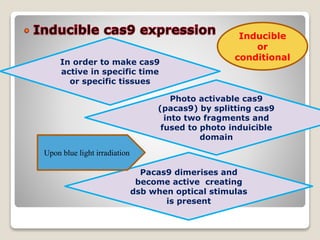
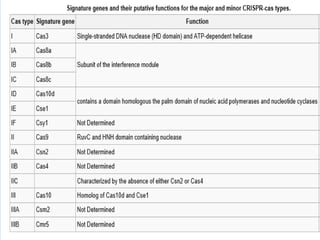
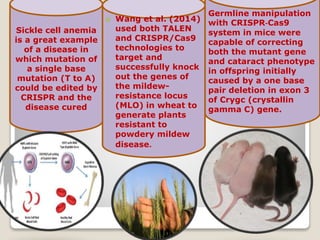
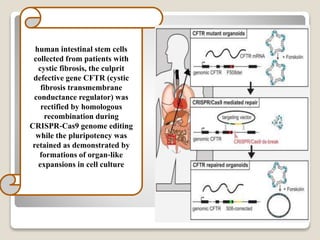
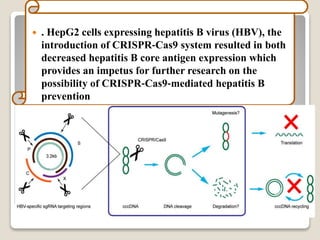
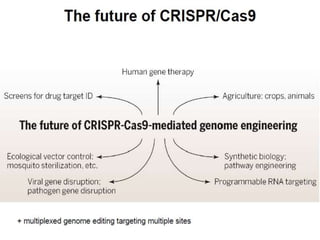
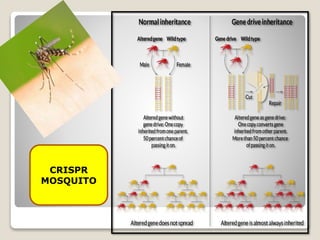
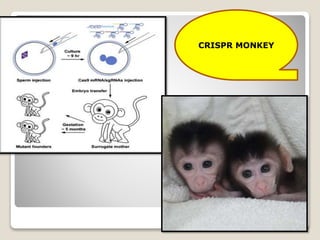
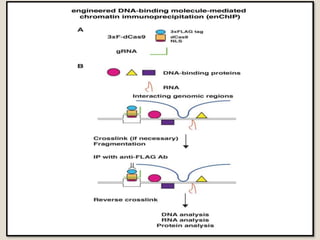
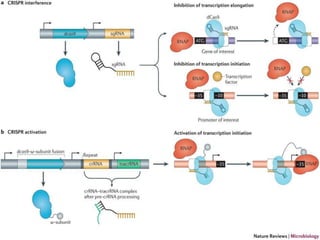
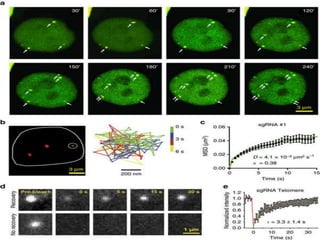
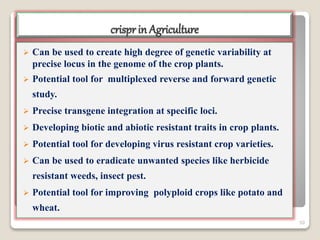
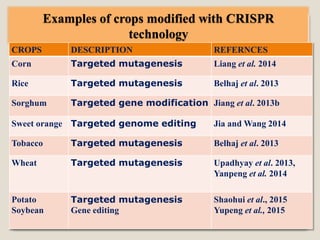
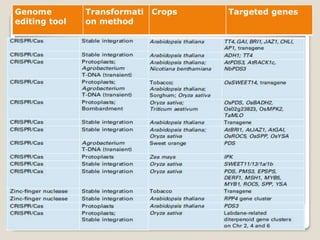
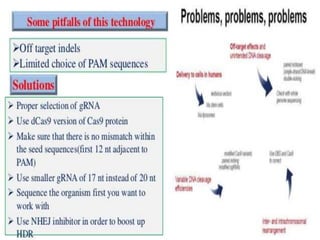
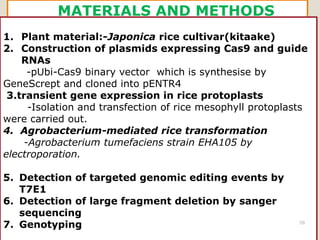
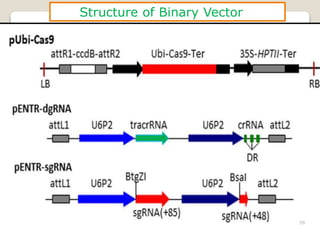
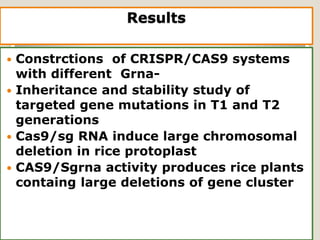
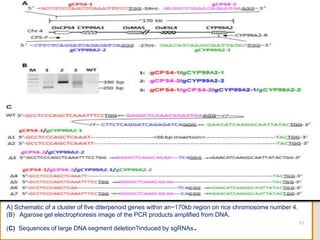
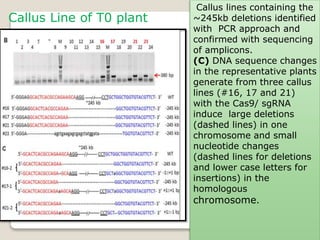
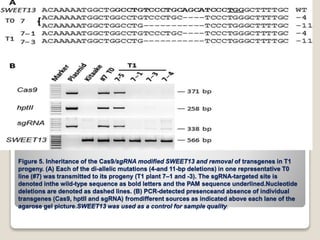
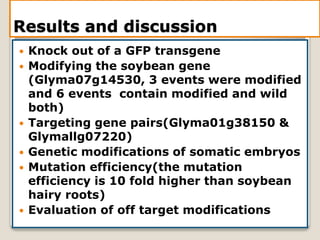
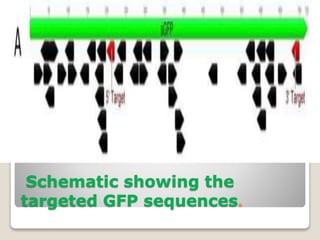
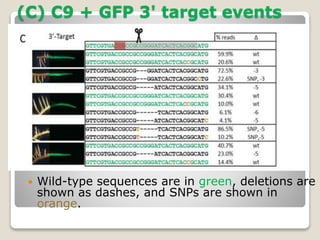
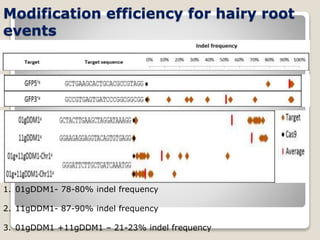
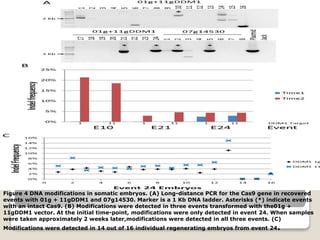
This document discusses the CRISPR-Cas9 genome editing technique. It begins with an overview of genome editing and provides a brief history. It then focuses on explaining CRISPR-Cas9, including its key components, how it was discovered as a natural bacterial immune system, and how it functions as a genomic tool. The document outlines the general CRISPR-Cas9 protocol and recent advances in the technique. It discusses applications in agriculture and for diseases. It also touches on advantages and limitations, as well as ethical issues. Two case studies are provided that demonstrate using CRISPR-Cas9 to modify genes in rice plants.