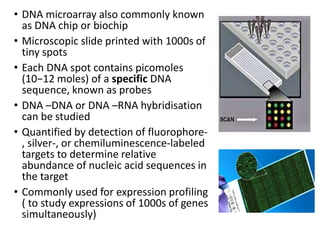
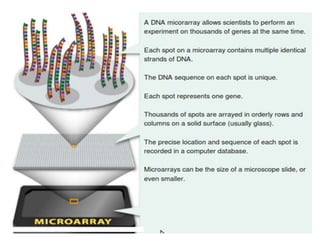
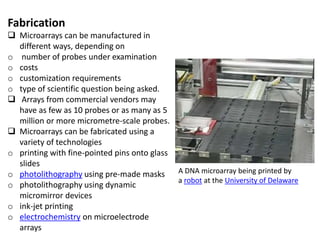
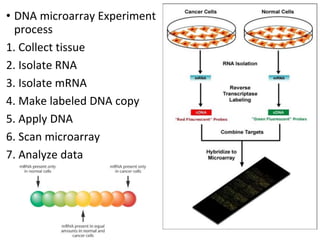
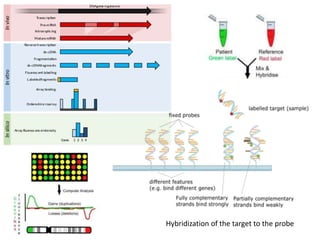
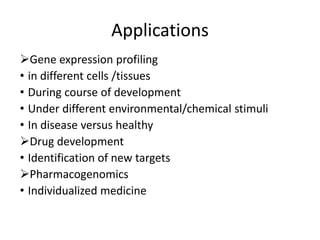
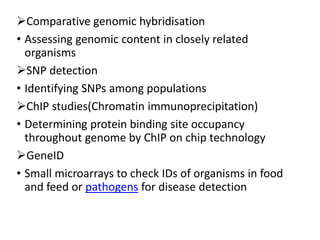
The document provides an introduction to DNA microarrays, detailing their composition, fabrication methods, and principles, including hybridization and quantitation of nucleic acid sequences. It discusses various applications such as gene expression profiling, drug development, pharmacogenomics, and comparative genomic hybridization, highlighting their significance in biomedical and clinical research. The advantages of microarrays include comprehensive genomic-wide information at relatively low costs and the ability to monitor gene expression patterns under different conditions.