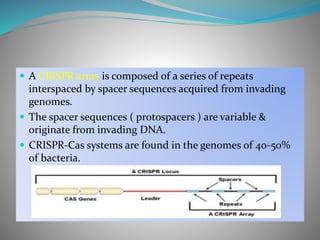
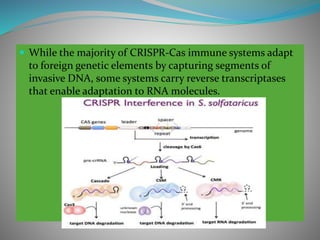
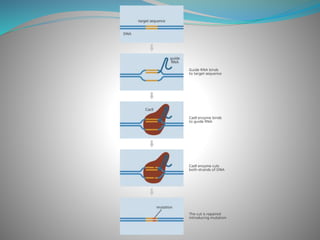
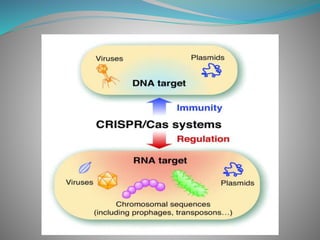
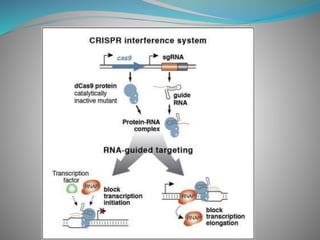
The document summarizes the CRISPR-Cas immune system found in bacteria and archaea. It has three main stages: adaptation, expression, and interference. The CRISPR-Cas9 system in particular allows for genome editing by creating targeted double-strand breaks in DNA directed by a guide RNA. This system is being used for genetic engineering in various organisms. The Cas9 endonuclease contains two nuclease domains that together cleave the target DNA.