1. The document discusses different types of cloning vectors including plasmids, bacteriophages, cosmids, and phagemids that can be used to clone foreign DNA.
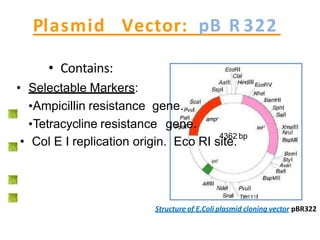
2. It provides details on the characteristics and components of common vectors like the pBR322 plasmid and lambda phage. Screening methods for identifying recombinant clones like blue/white selection and replica plating are also described.
3. Applications of genetic engineering using these vectors include producing recombinant proteins like insulin and hepatitis vaccines as well as studying gene function and identifying mutations associated with diseases.