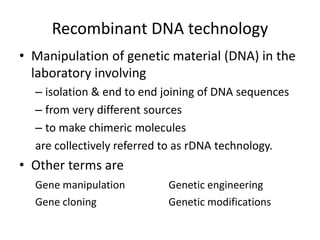
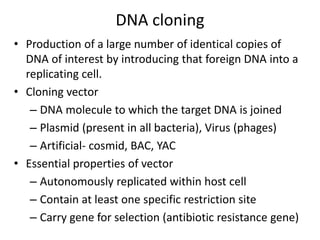
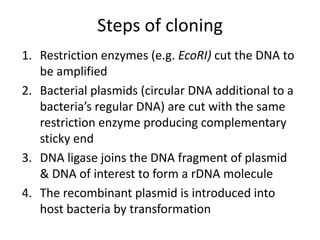
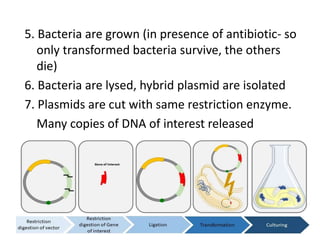
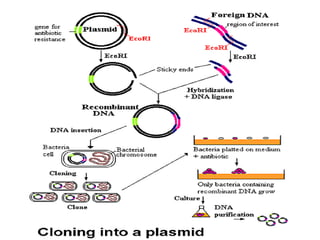
1. Recombinant DNA technology involves manipulating genetic material from different sources to create chimeric DNA molecules. This is done using various tools like restriction enzymes, DNA ligase, cloning, and gene transfer methods.
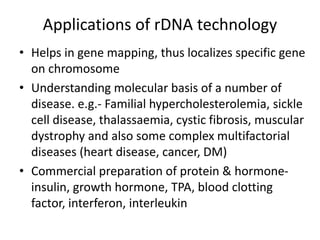
2. Medical biotechnology applies recombinant DNA technology to research and produce pharmaceuticals and diagnostic products to treat and prevent human diseases. This includes producing insulin, growth hormones, antibodies, and vaccines.
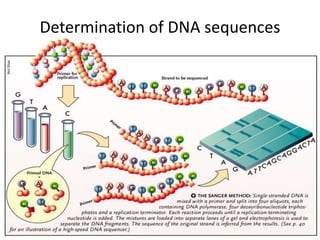
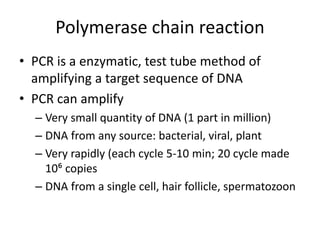
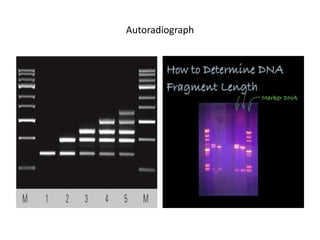
3. Techniques like PCR, blotting, hybridization, and DNA sequencing are used in recombinant DNA technology applications such as gene mapping, understanding disease mechanisms, DNA-based diagnostics, forensics, and gene therapy.


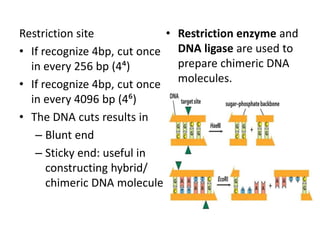
![Restriction enzyme
• Endonuclease
– Cut DNA at specific DNA sequences within the
molecule
– Into unique, short pieces in a sequence specific
manner (not randomly)
• Named after the bacterium from which they are
isolated. [EcoRI- from E. Coli]
• Each enzyme recognizes & cleaves a specific &
double stranded DNA sequence
– 4-7 bp long
– Palindrome: same sequence from 5΄3΄ direction](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-7-320.jpg)
![Library
A library- is a collection of recombinant clones
– Genomic library: prepared from total DNA of a cell
line or tissue
– cDNA library: only expressed DNA [cDNA copies of
mRNA] in a tissue](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-9-320.jpg)
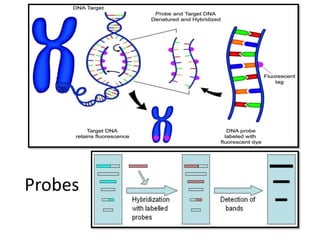
![Probes
• Probe is a molecule used to detect the presence of
a specific fragment of a DNA or RNA.
• Short piece (15-20 NT) of ssDNA or RNA, labeled
with a radioisotope [³²P]or fluorescent dye
• Complementary to a sequence of the DNA of
interest/ target DNA
• Used to screen a library for a complementary
sequence in the coding region of the gene.
• If the sequence match exactly, probes will hybridize](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-14-320.jpg)
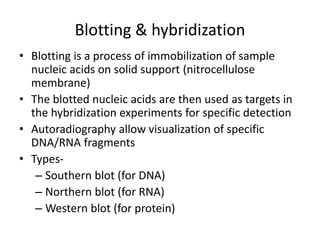
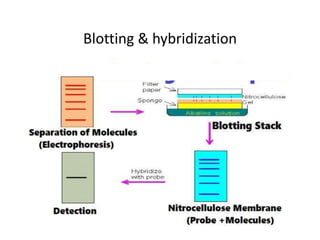
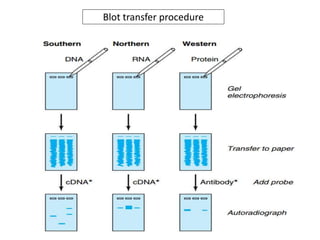
![1. Southern blot: A method of transferring DNA
from an agarose gel to nitrocellulose filter, on
which the DNA can be detected by a suitable
probe [cDNA or RNA]
2. Northern blot: A method of transferring RNA
from an agarose gel to nitrocellulose filter, on
which the RNA can be detected by a suitable
probe [cDNA or RNA]
3. Western blot: A method of transferring protein
to a nitrocellulose filter, on which the protein
can be detected by a suitable probe [antobody]](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-18-320.jpg)
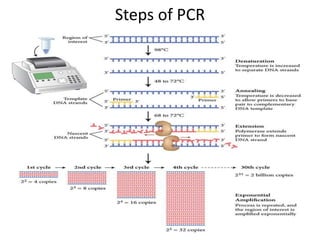
![Steps of PCR
• Use DNA polymerase from Thermus Aquaticus [TaqP].
Heat stable, so not denatured at 70-80⁰C.
• Primer construction [20-35 NT] according to flanking
sequence
1. Denaturation of DNA [94⁰C]
2. Annealing of primers to ssDNA [54⁰C]
3. Chain extension by DNA-P [72⁰C]
New dsDNA molecules can be denatured & copied
repeatedly](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-22-320.jpg)
![Uses of PCR
• Forensic analysis of DNA sample
• To detect infectious agent [latent virus-HIV]
• To make prenatal genetic diagnosis
• To detect allelic polymorphism
• To establish precise tissue types for transplants
• To study evolution, using DNA from archeological
samples
• Quantitate RNA analysis [RT-PCR]
• To score in vivo protein DNA occupancy using
chromatin immuno-precipitation assay to facilitate NGS
[next generation occupancy]
New uses are developed every year.](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-25-320.jpg)
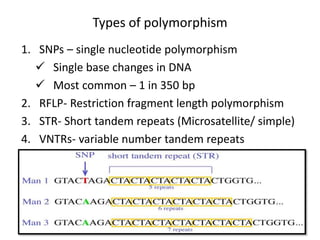
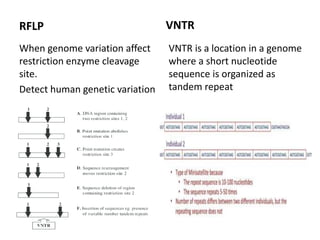
![Polymorphism
• Change in genotype that does not affect phenotype or
may change phenotype that is harmless.
• Primarily occur in non-coding sequence [only 2%
genome encode protein in human]
• Genome of any 2 unrelated people are 99.5% identical
Important tool in
– genome mapping
– localization of genes for genetic disorders
– determination the risk for disease
– paternity testing, criminal identification](https://image.slidesharecdn.com/rdnatechnology-200223123158/85/Recombinant-DNA-Technology-26-320.jpg)