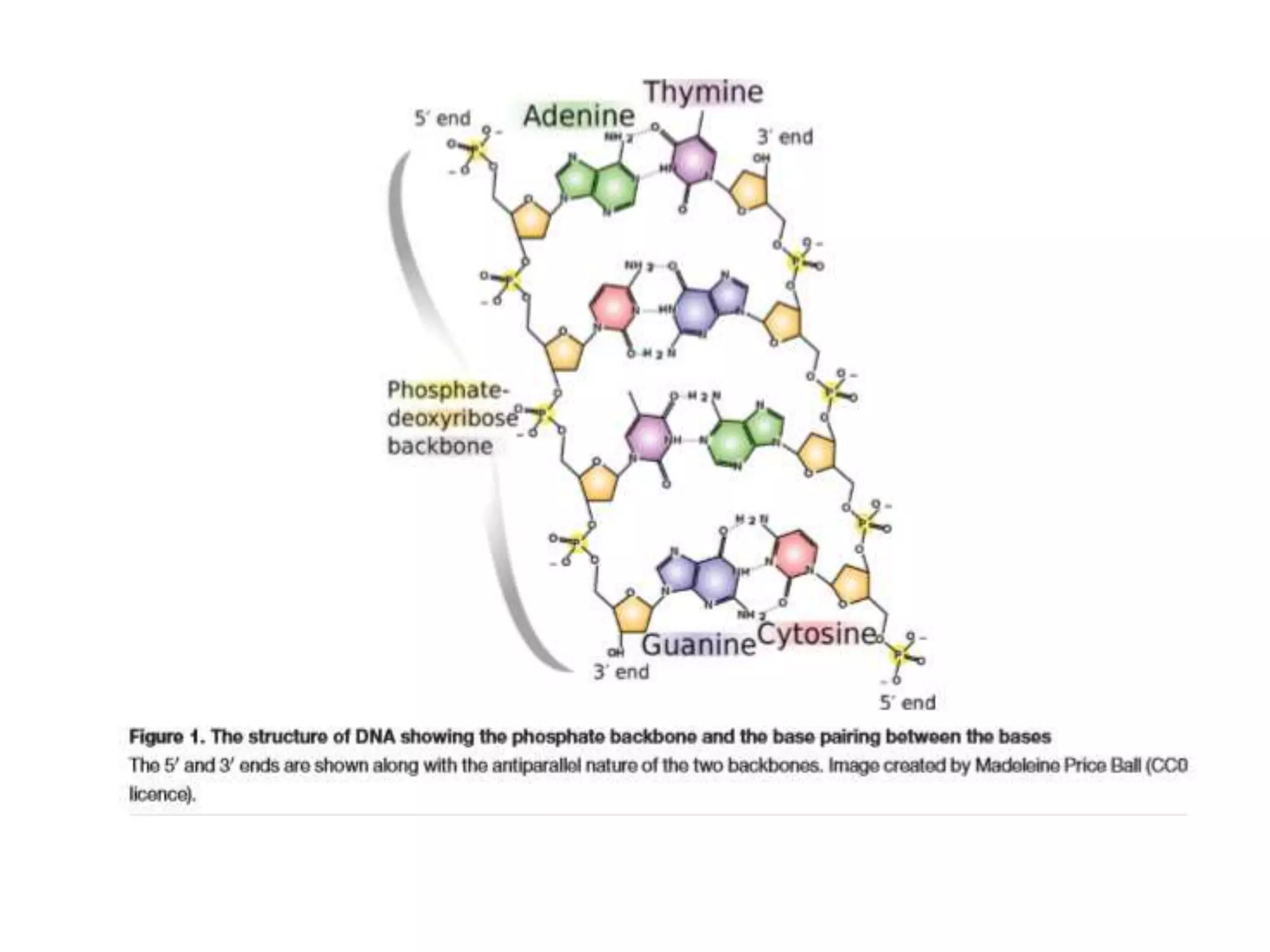
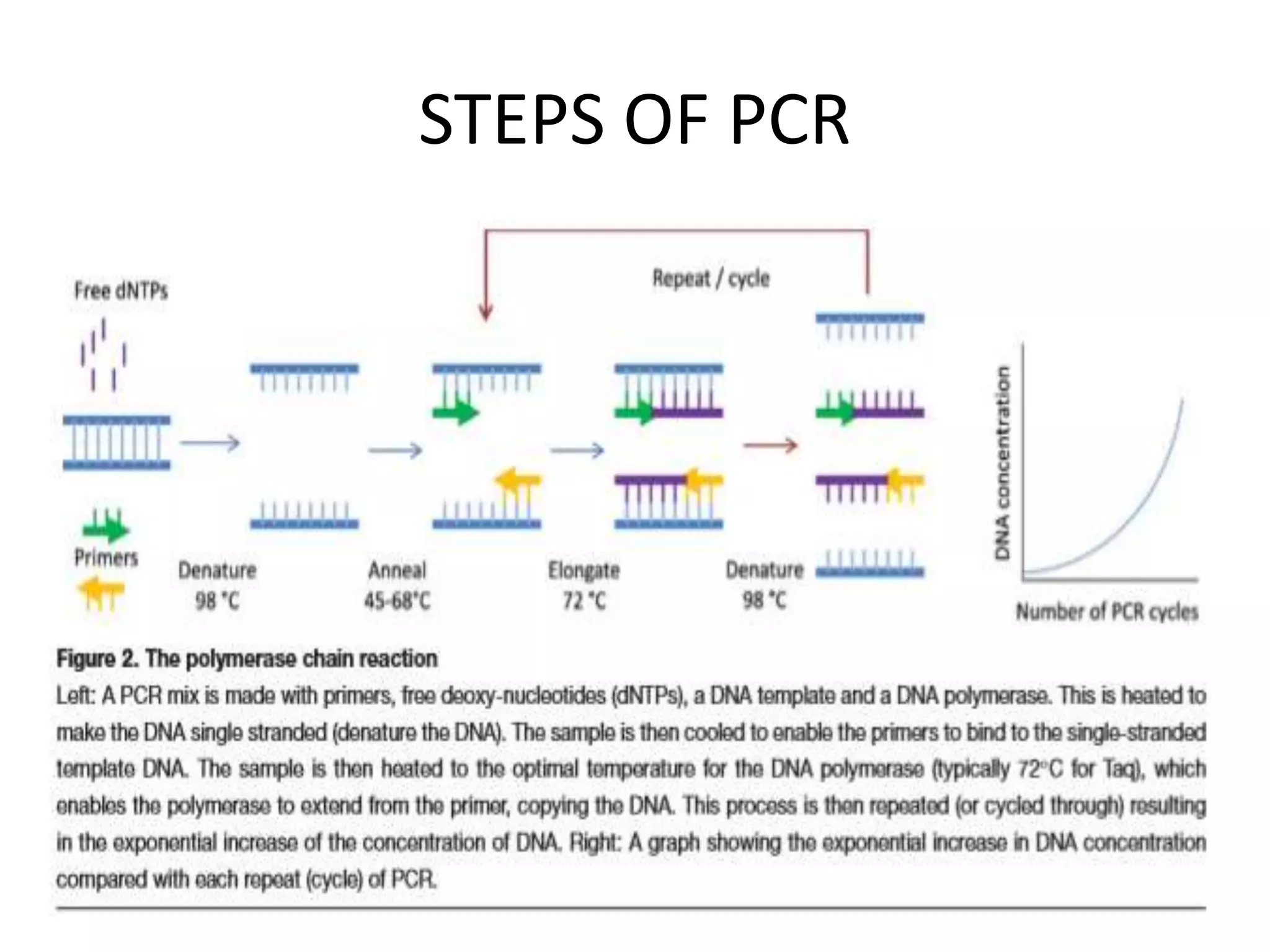
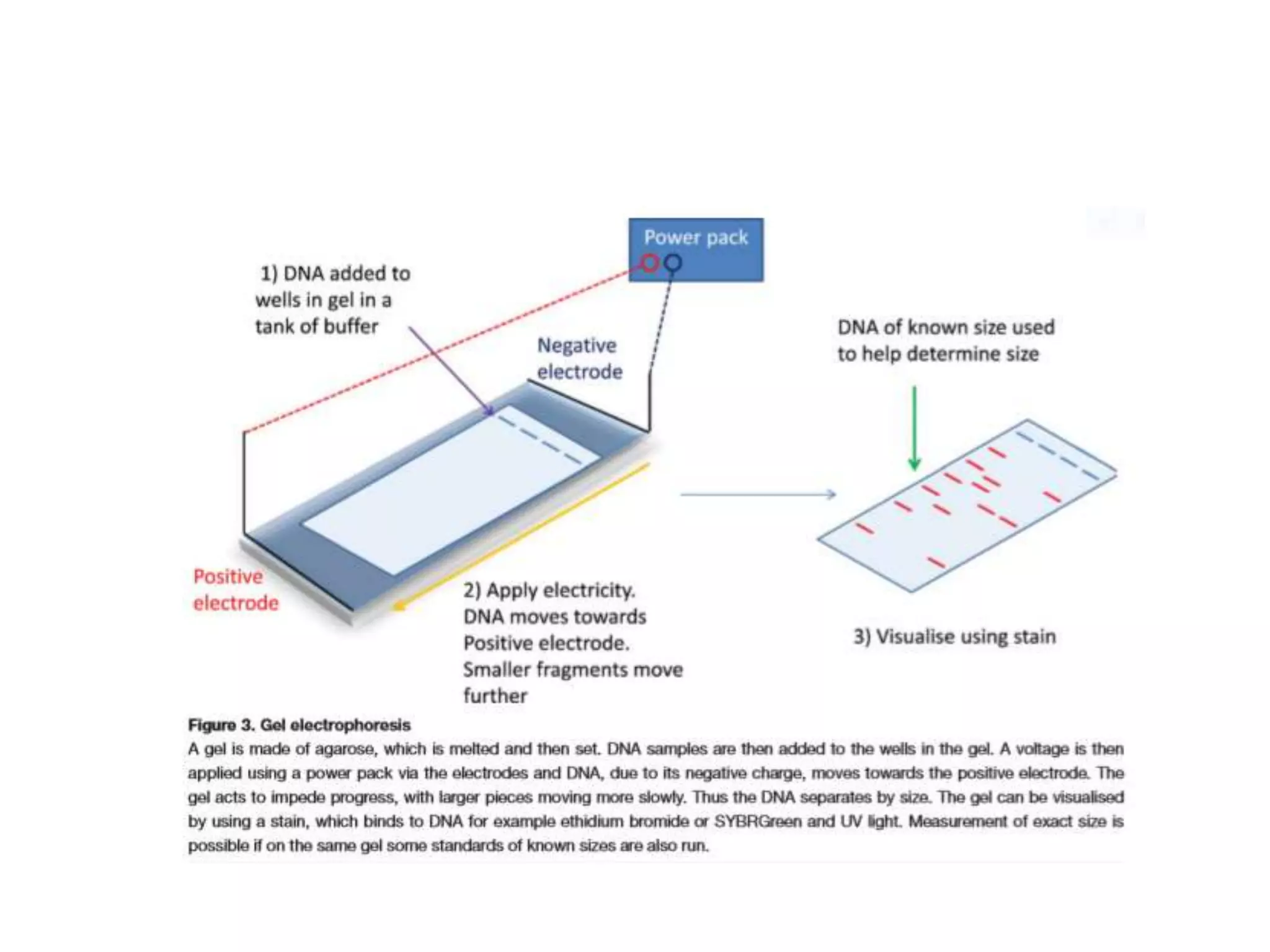
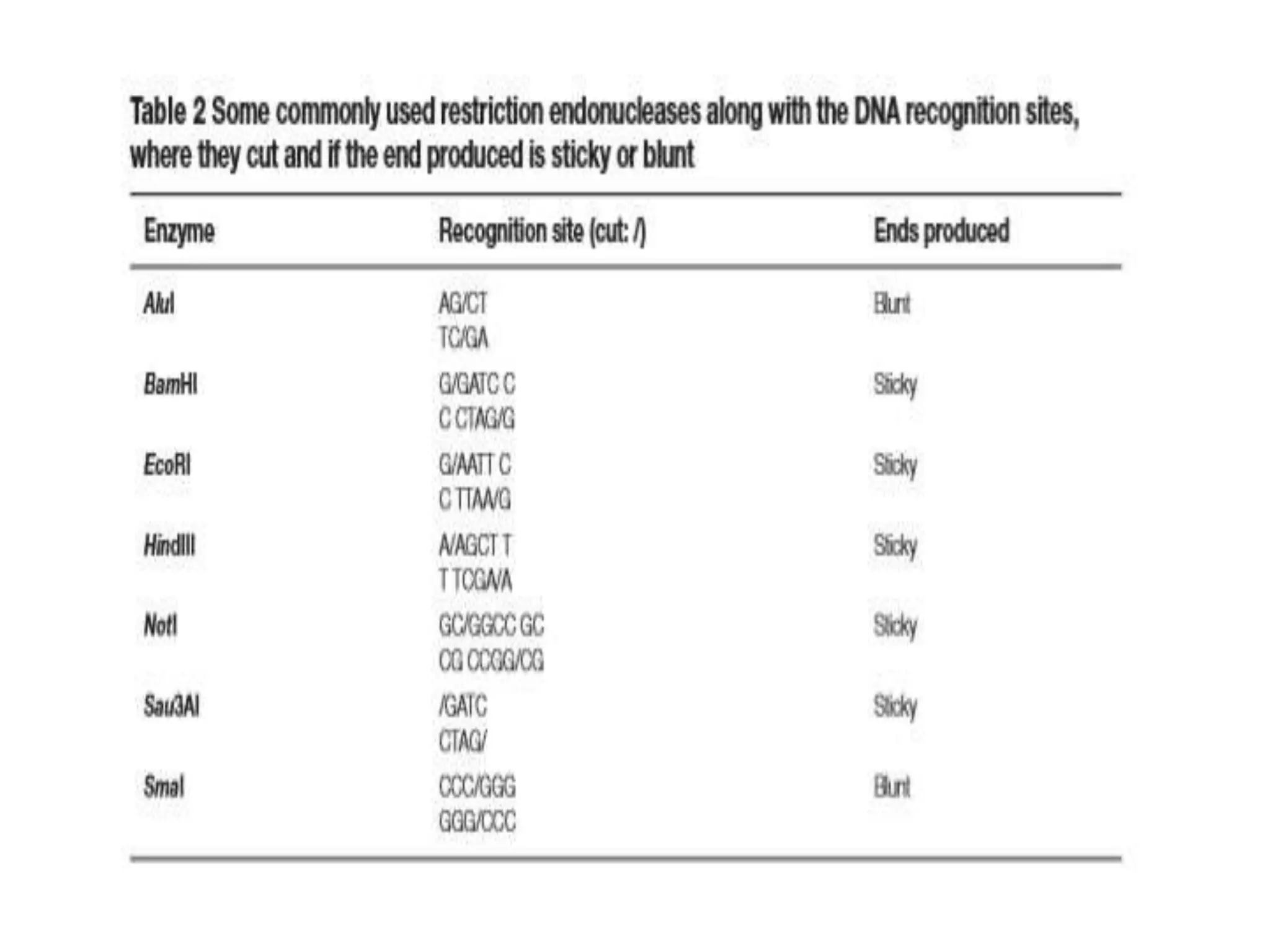
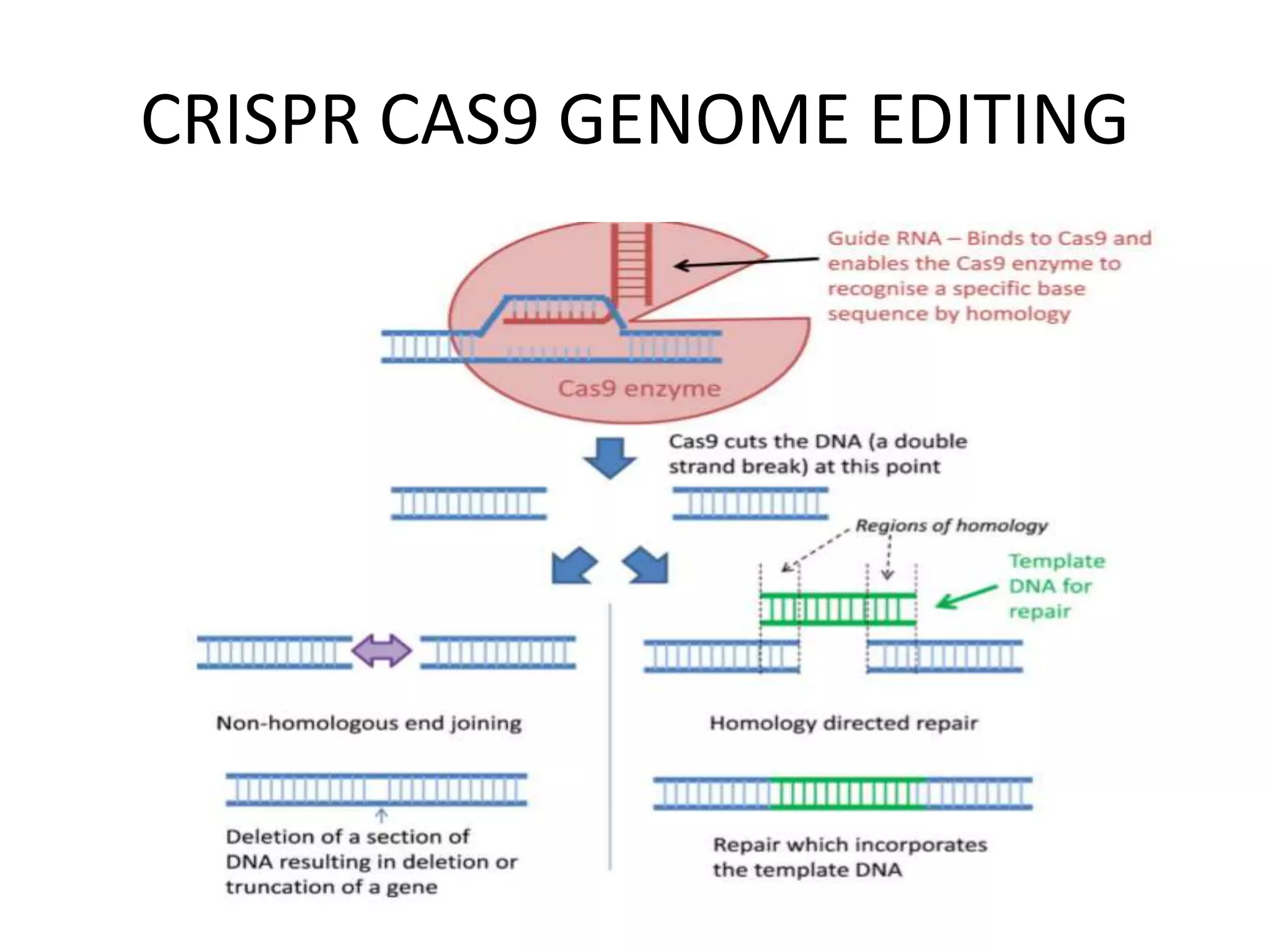
The document provides an overview of recombinant DNA technology and DNA sequencing, detailing the structure and function of DNA, methods for DNA isolation, amplification through polymerase chain reaction (PCR), and DNA manipulation techniques. It emphasizes the importance of vectors for storing DNA and methods such as restriction endonucleases and DNA ligase for joining DNA fragments. Additionally, the document touches on CRISPR-Cas9 as a genome editing tool.