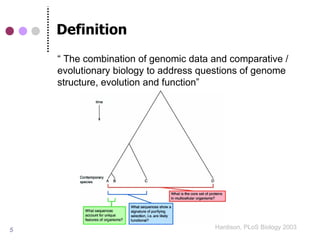
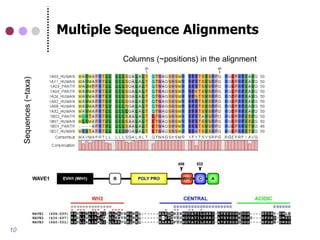
The document discusses comparative genomics in eukaryotes, emphasizing the study of genome relationships due to common ancestry. It highlights the value of cross-species comparisons for understanding genome conservation, variation, and functional elements, as well as the importance of accurately interpreting homology in sequence analysis. The presentation includes information on genome databases and the significance of conserved gene structures and regulatory elements.