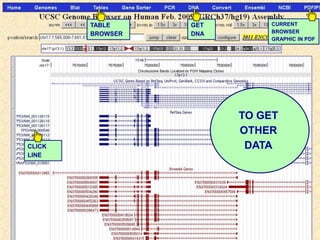
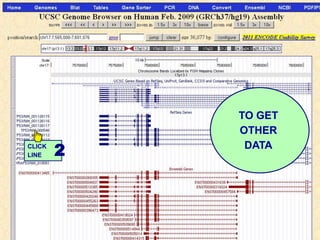
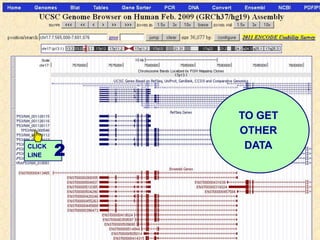
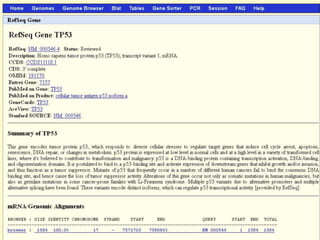
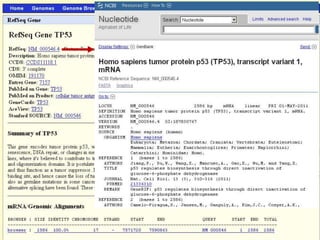
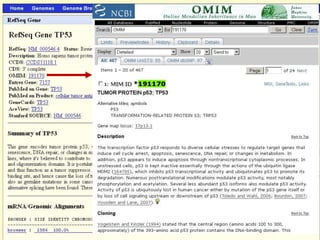
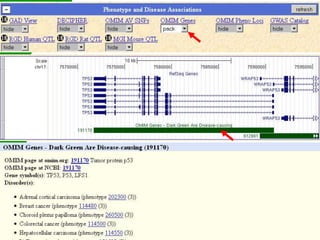
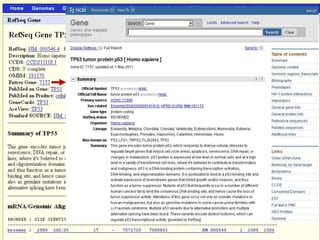
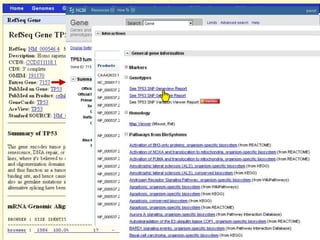
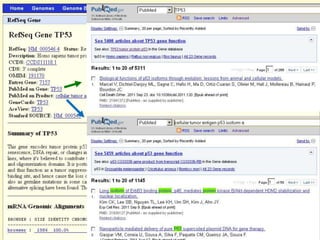
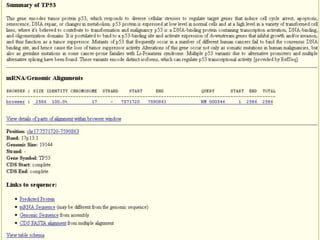
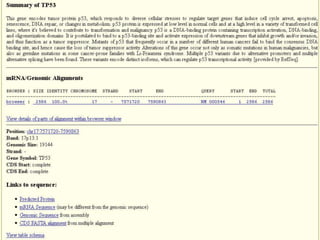
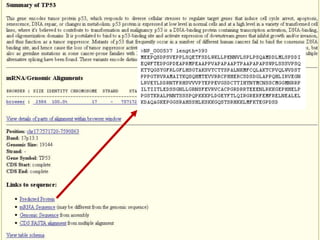
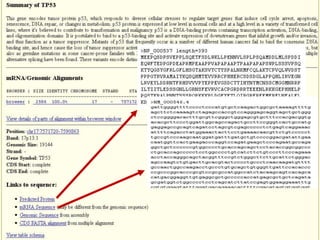
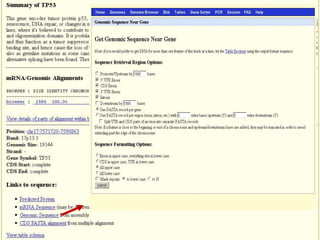
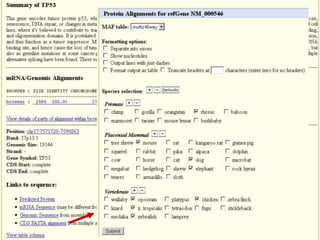
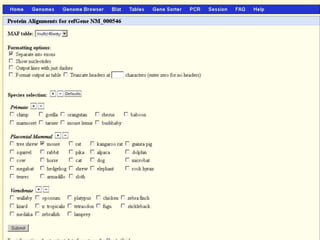
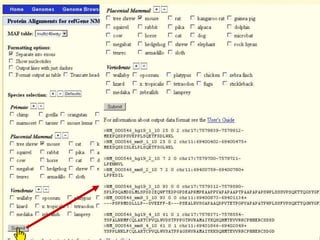
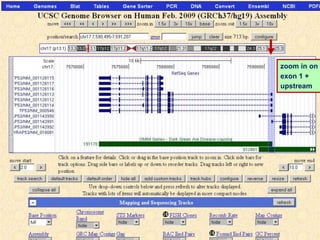
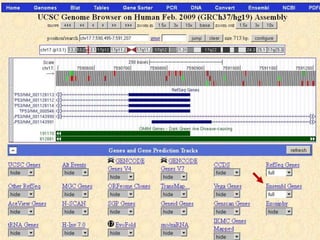
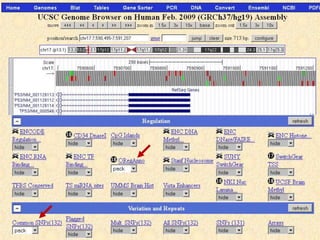
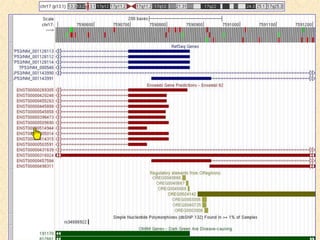
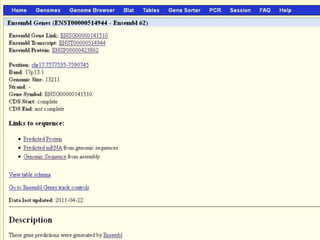
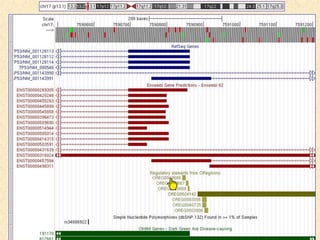
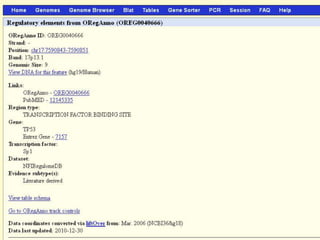
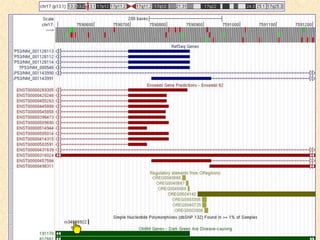
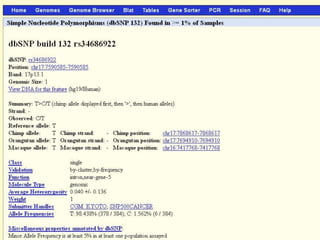
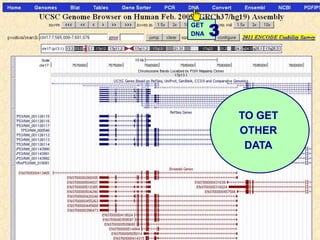
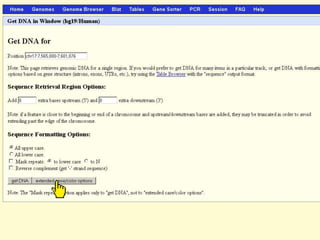
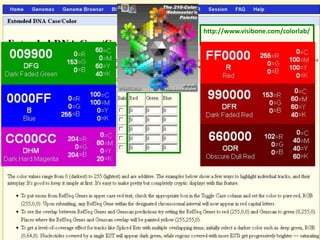
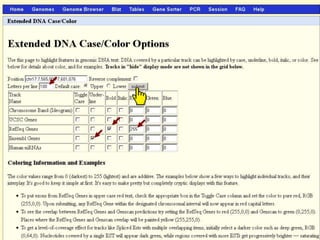
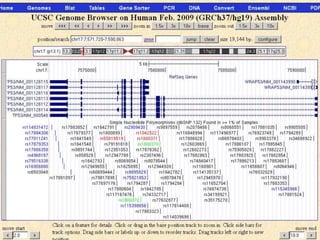
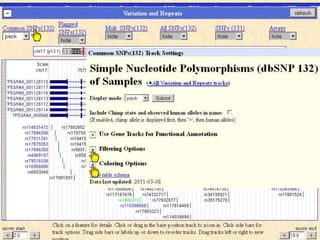
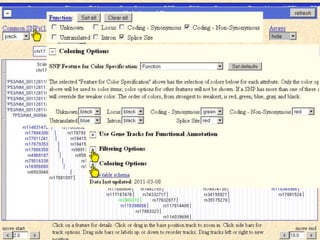
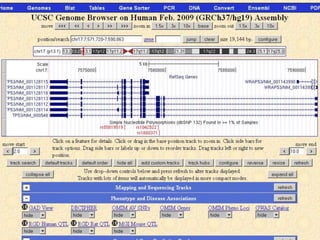
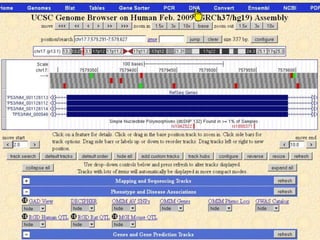
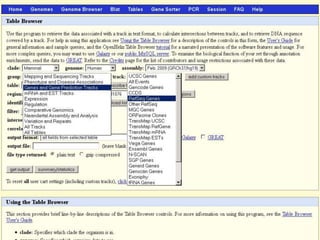
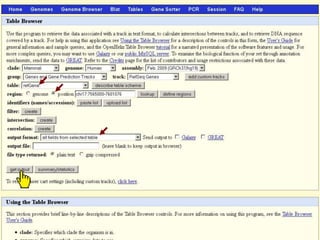
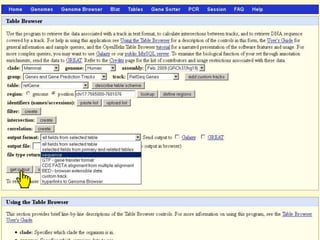
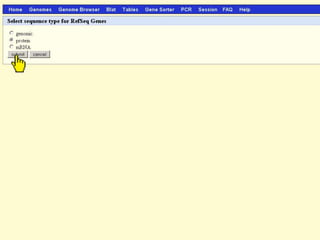
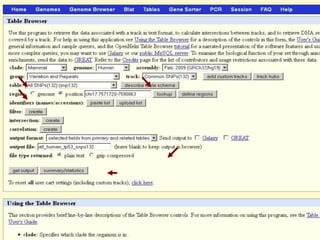
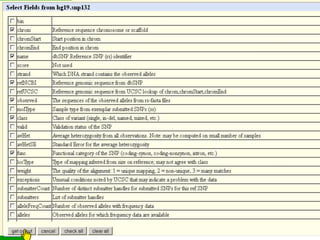
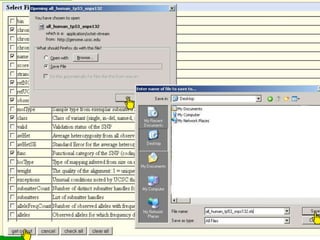
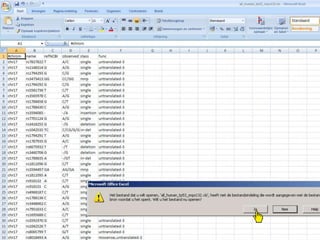
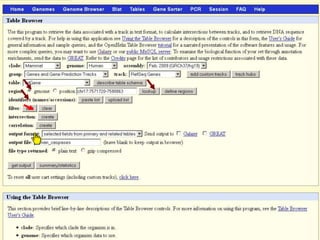
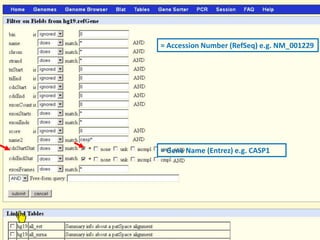
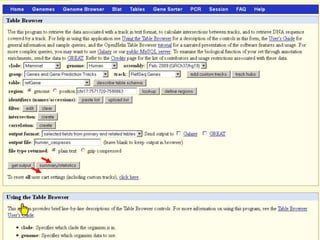
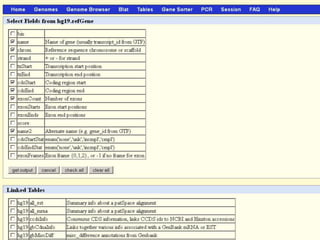
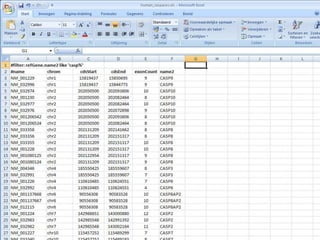
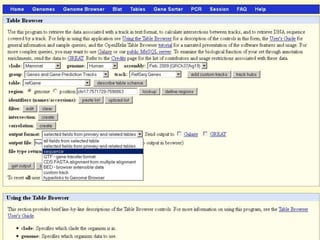
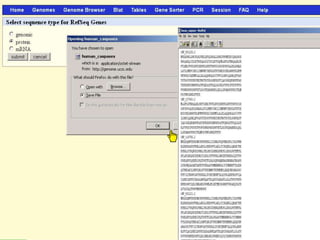
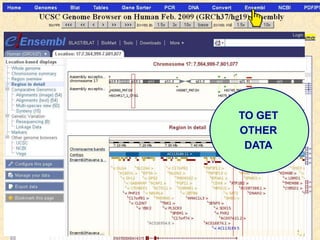
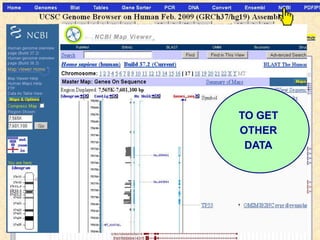
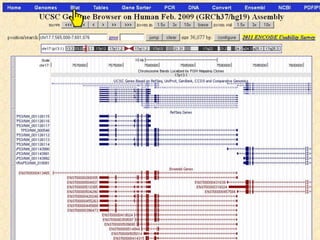
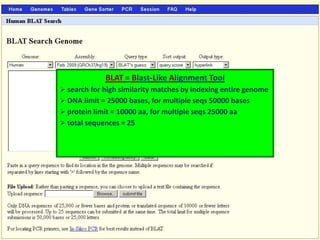
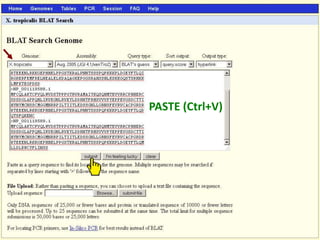
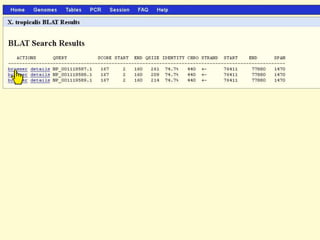
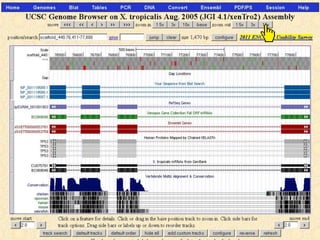
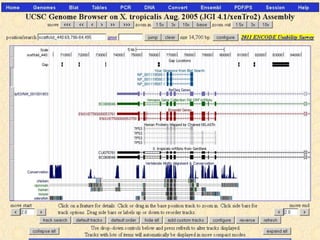
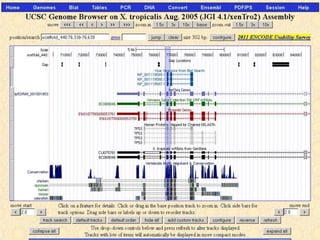
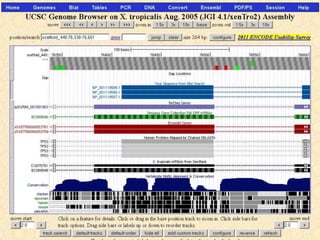
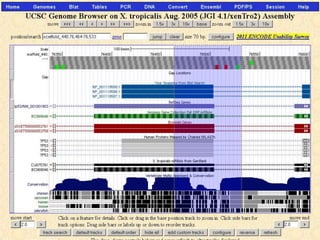
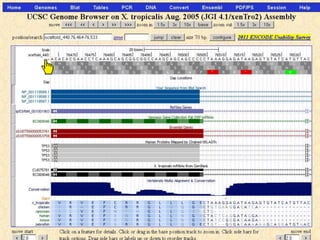
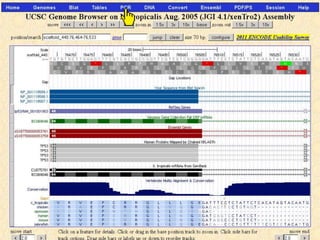
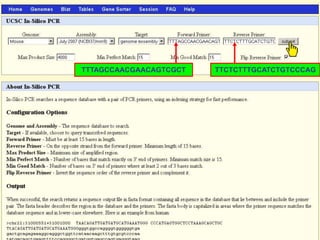
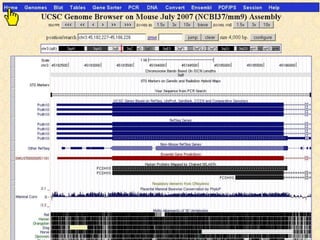
The document describes various resources and tools available for genomic research, including GenBank, RefSeq, and Ensembl for gene annotation. It includes exercises for retrieving gene sequences, analyzing SNPs, and using alignment tools like BLAT to find orthologs. Additionally, it provides guidelines for saving results in different formats for further analysis.