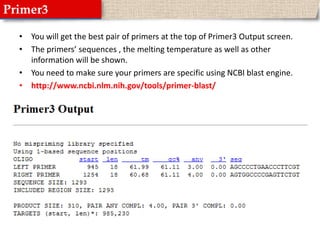
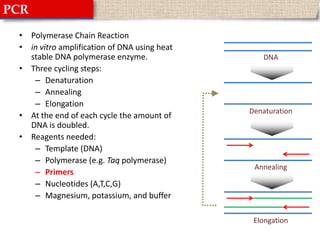
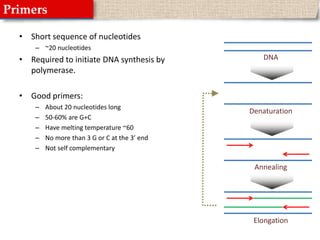
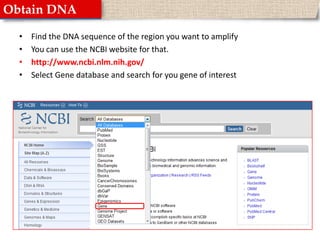
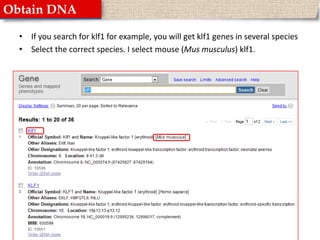
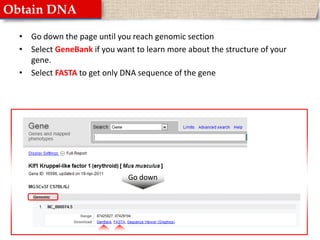
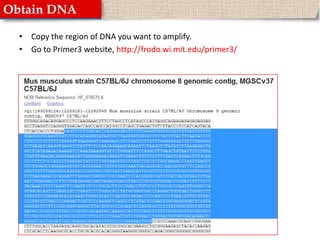
This document provides instructions for designing primers for PCR using the Primer3 website. It explains that PCR involves cycling between DNA denaturation, annealing of primers, and elongation to amplify a target DNA sequence. Good primers are around 20 nucleotides long with a GC percentage of 50-60% and melting temperature around 60°C. The document guides obtaining a target DNA sequence from NCBI and using Primer3 to design primers, specifying amplicon size and target region. It recommends checking primer specificity using NCBI BLAST.

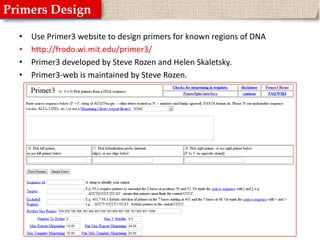
![Primer3Paste the DNA sequence into the provided spaceTell the website where do you want you primers to be by adding “[“ and “]” around the sequence of interst.Tell the website what is the size of amplicon you would like to see.Hit “Pick Primers”](https://image.slidesharecdn.com/primerdesign-110504183521-phpapp01/85/Primer-design-10-320.jpg)