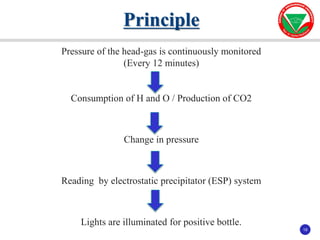
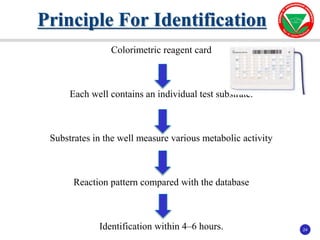
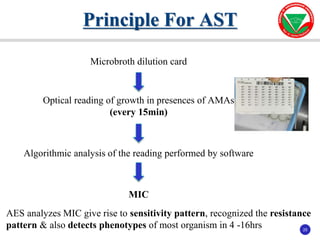
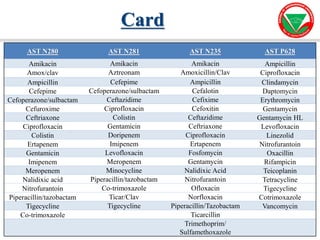
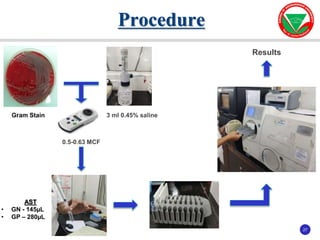
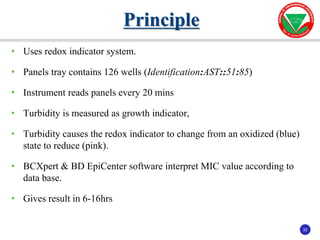
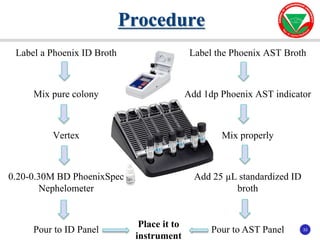
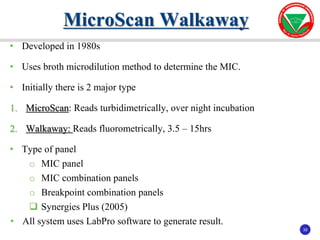
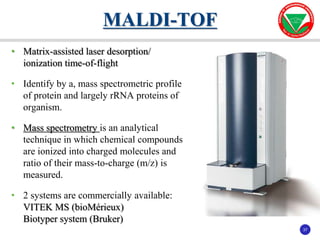
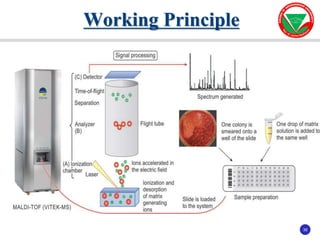
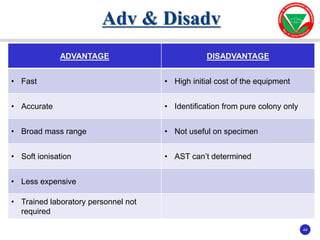
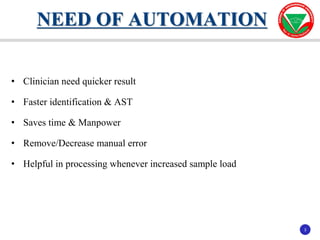
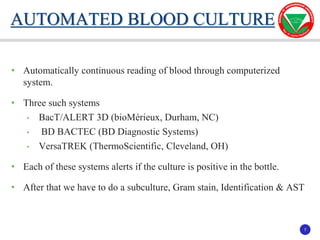
The document summarizes various automated systems used in bacteriology. It discusses automated blood culture systems like BacT/ALERT 3D, BD BACTEC, and VersaTREK which continuously monitor blood cultures. It also describes automated identification and antimicrobial susceptibility testing systems like Vitek, Phoenix, and MicroScan Walkaway. Finally, it provides an overview of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) which uses the mass spectra of proteins to rapidly identify bacteria and yeast.







![10
10
Media
AEROBIC CULTURE ANAEROBIC
MEDIA
MYCOBACTERIUM
MEDIA
BactAlert FA
Plus
BactAlert PA
Plus
BactAlert FN
Plus
BactAlert MB Process
SAMPLE
Blood & Sterile
Fluid
Blood Blood & Sterile
Fluid
All Specimens Except
Whole Blood
BOTTLE
CONTAINS
• 30 ml of Broth [BHI and TSB]
• Soduim poly-an-ethol-sulfonate
• Nutrients, Amino acids, Carbohydrate substrates
• APB (Adsorbent Polymeric Beads)
In anaerobic media 40ml
• 10 ml Middle brook
7H9
• Pancreatic Digest of
casein.
• Bovin serum albumin.
• Catalase in purified
water
SAMPLE
VOLUME
5 to 10ml 1 to 4ml 1 to 10ml 0.5 ml after
decontamination](https://image.slidesharecdn.com/automationinbacteriologydr-210902105916/85/Automation-in-bacteriology-dr-sumesh-10-320.jpg)