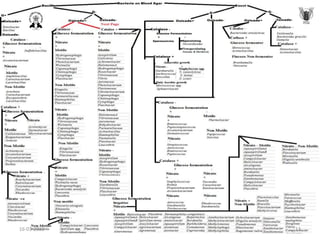
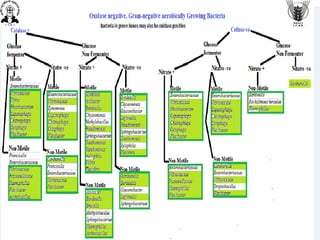
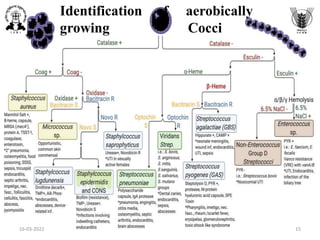
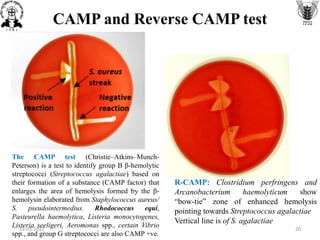
The document discusses conventional methods for identifying and characterizing pathogenic bacteria, emphasizing the limitations of automated systems and the importance of pure cultures for accurate identification. It outlines various identification techniques, including biochemical tests, culturing methods, and advanced molecular methods, while detailing specific media used for selective enrichment and plating of different bacteria. Additionally, it highlights essential tests and assays required to determine the virulence potential of bacterial isolates.




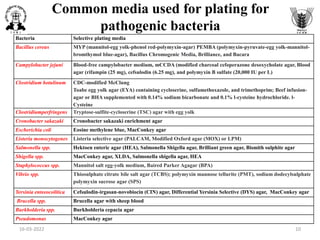
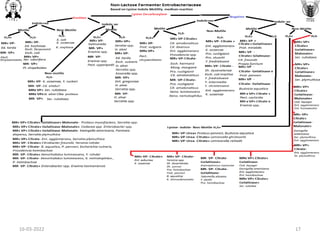
![Common media used for selective enrichment
of pathogenic bacteria
Bacteria Selective Enrichment broth
Bacillus cereus BPW heated to pasteurize after inoculation
Campylobacter jejuni Selective enrichment medium (ATB)
Clostridium botulinum TPGY broth media, Egg meat medium fortified with 1% additions of yeast extract, ammonium sulfate,
and glucose (FEM medium)
Clostridiumperfringens Thioglycollate medium (fluid) , iron-milk medium
Cronobacter sakazaki Cronobacter sakazaki enrichment broth
Escherichia coli Enterobacteriaceae enrichment broth, MacConkey broth
Listeria monocytogenes Al-Zoreky-Sandine Listeria medium [ASLM] ; University of Vermont Medium (UVM) containing
acriflavin and naladixic acid; Fraser broth with acriflavin, naladixic acid and esculin; Listeria
Enrichment broth (LEB, FDA BAM formulation) containing the selective agents acriflavin, naladixic
acid and the antifungal agent cycloheximide
Salmonella spp. Tetrathionate broth Rappaport Vassiliadis medium, Selenite broth
Shigella spp. Shigella broth with 0.5mg/L novobiocin
Staphylococcus spp. BHI with 10% NaCL
Vibrio spp. Alkaline peptone water (APW), Glucose salt teepol (or sodium dodecylsulphate) broth (GSTB)
Yersinia enteeocolitica ICT or TTC Broth (contains irgasan or triclosan)
Brucella spp. BHI broth
Burkholderia spp. Burkholderia broth
Pseudomonas Dettol broth, BHI broth
16-03-2022 9](https://image.slidesharecdn.com/conventionalmethodsforidentificationandcharacterizationofpathogenicbacteria-220312061113/85/Conventional-methods-for-identification-and-characterization-of-pathogenic-bacteria-9-320.jpg)