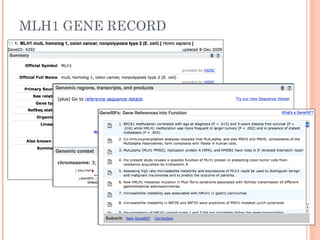
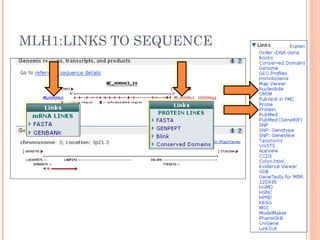
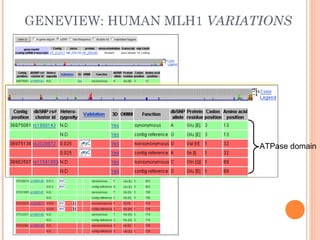
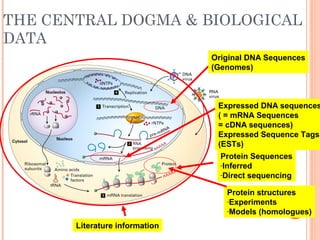
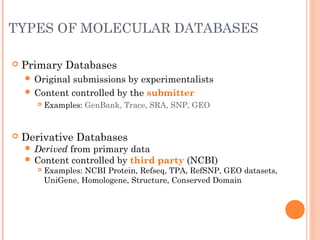
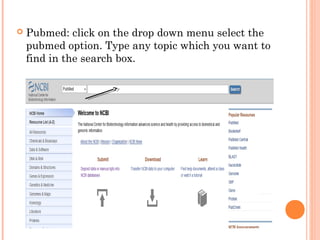
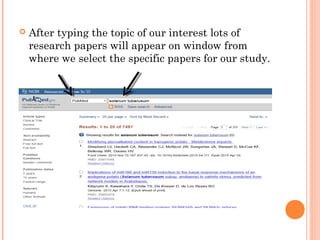
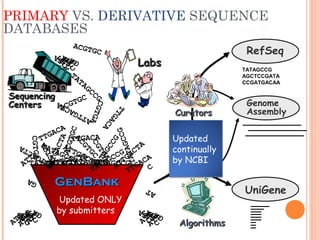
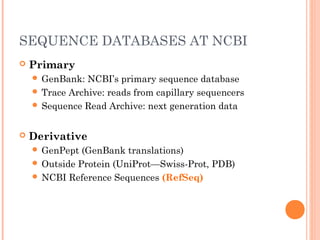
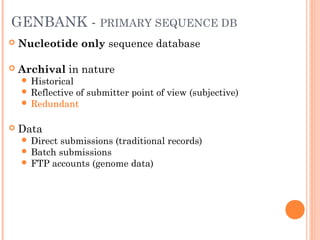
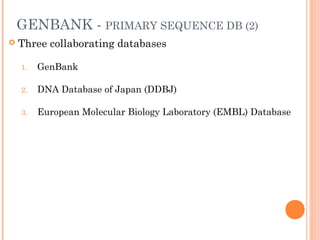
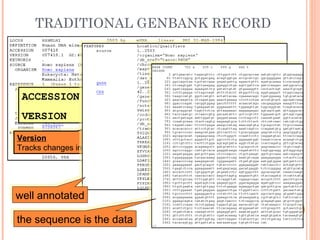
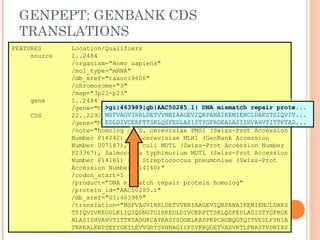
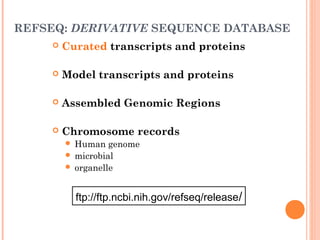
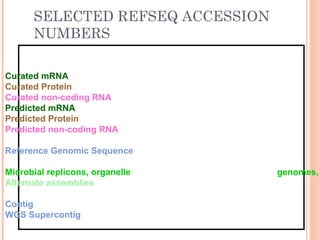
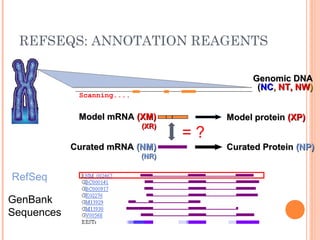
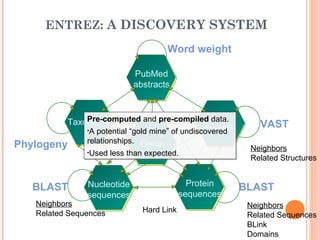
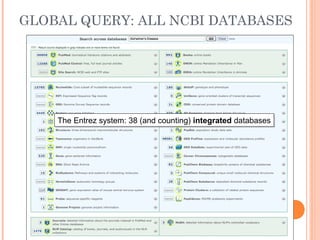
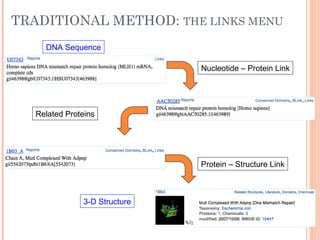
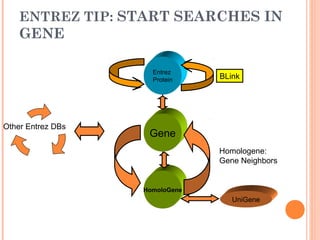
This document provides an introduction to biological databases. It discusses what databases are and features of an ideal database. It describes the relationships between primary sequence databases like GenBank that contain original submissions, and derived databases like RefSeq that are curated by NCBI. Key databases at NCBI are described, including GenBank, RefSeq, and Entrez, which allows integrated searching across multiple databases. The benefits of data integration through linking related information are highlighted.














![PRECISE RESULTS
MLH1[Gene Name] AND Human[Organism]MLH1[Gene Name] AND Human[Organism]](https://image.slidesharecdn.com/biologicaldatabases-151208123300-lva1-app6892/85/Biological-databases-34-320.jpg)