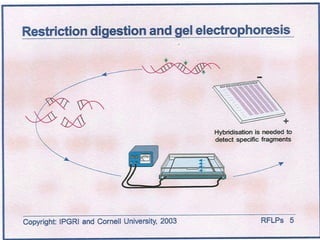
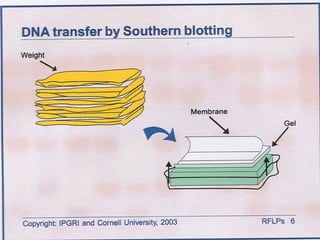
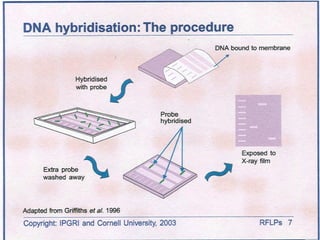
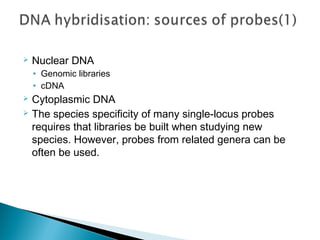
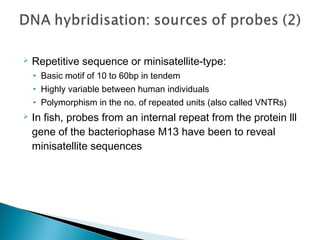
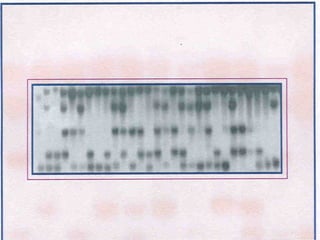
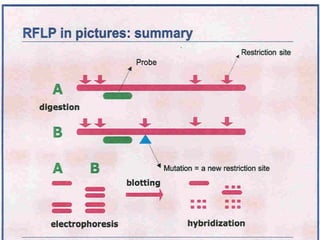
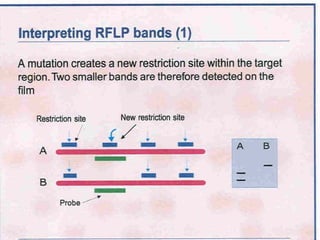
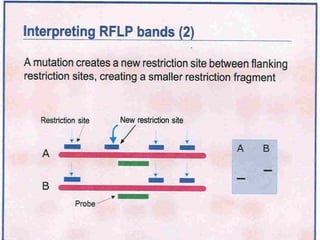
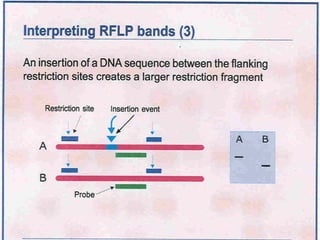
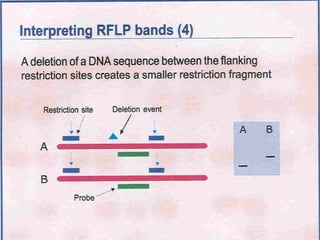
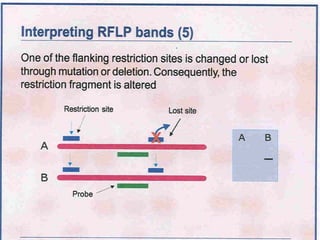
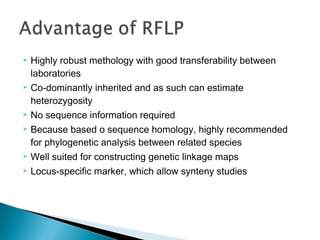
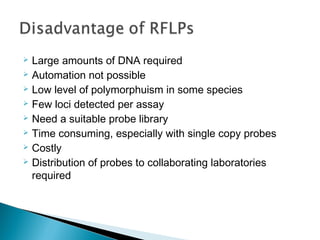
This document discusses the RFLP (restriction fragment length polymorphism) technology. It describes the key steps in RFLP which are isolating DNA from samples, digesting the DNA with restriction enzymes, separating fragments via gel electrophoresis, transferring DNA to membranes, and detecting bands through hybridization with probes. The document outlines the equipment needed and discusses advantages like being robust and able to estimate heterozygosity, as well as disadvantages like being time-consuming and requiring large DNA amounts. Finally, it presents applications of RFLP in areas like genetic diversity analysis and gene mapping.