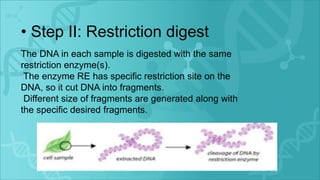
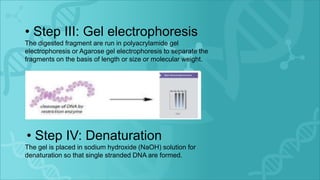
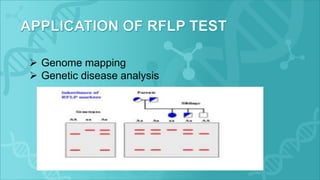
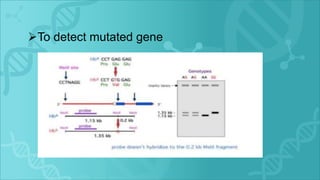
RFLP is a technique that uses restriction enzymes to cut DNA into fragments at specific nucleotide sequences. These fragments are then separated by gel electrophoresis and analyzed by hybridization with labeled probes. Differences in the fragment patterns between individuals or strains can be used for applications like genome mapping, disease analysis, and criminal investigations. RFLP provides semi-dominant markers but requires high quality DNA and is a long, labor-intensive process.