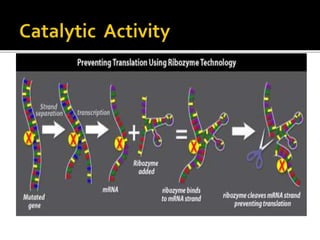
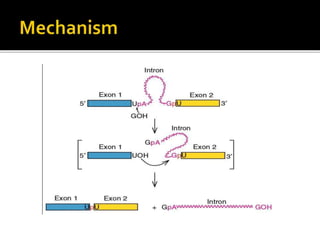
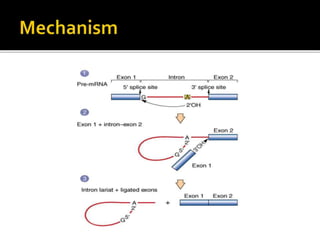
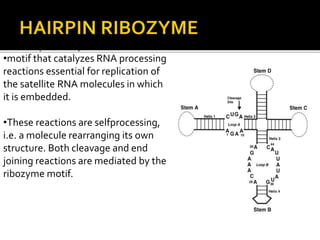
Ribozymes are RNA molecules that can catalyze biochemical reactions like protein enzymes. The first ribozyme was discovered in 1980. There are two main classes of natural ribozymes - self-cleaving ribozymes like hammerhead and hairpin ribozymes, and self-splicing ribozymes like group I and group II introns and RNase P. Ribozymes are being investigated for their potential in gene therapy applications by specifically cleaving target mRNA molecules.