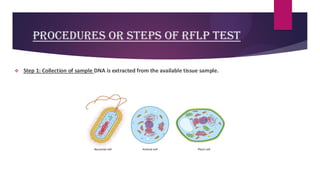
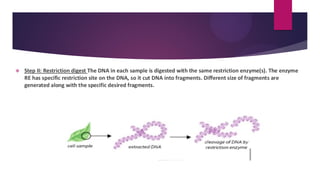
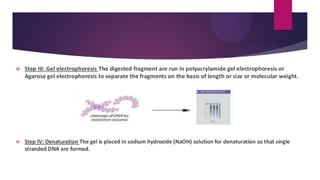
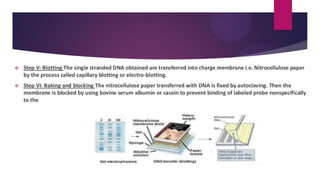
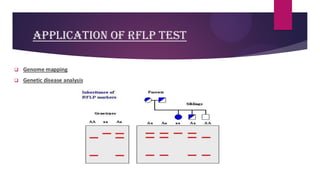
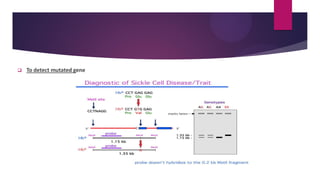
This document discusses restriction fragment length polymorphism (RFLP), a technique used for DNA fragment separation and identification. It outlines the principles, procedural steps, applications, advantages, and disadvantages of RFLP, emphasizing its roles in genome mapping, genetic disease analysis, and criminal investigations. Key considerations include the need for high-quality DNA, the complexity and labor intensity of the methodology, and the high costs associated with its use.