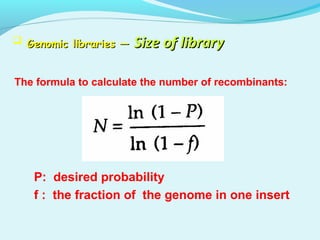
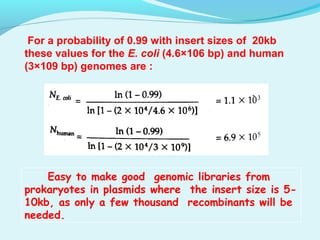
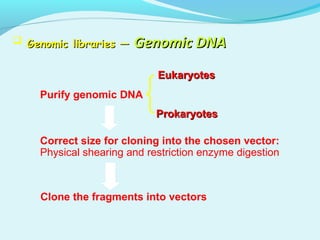
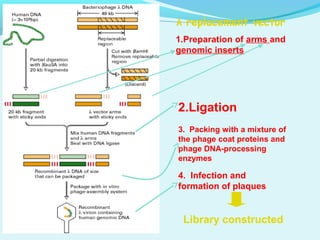
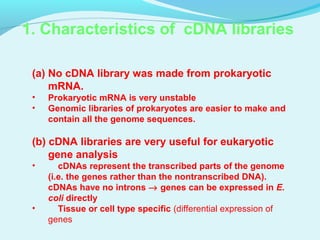
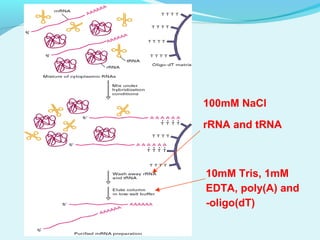
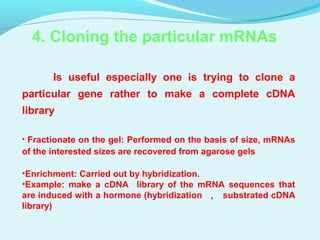
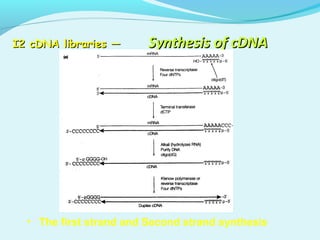
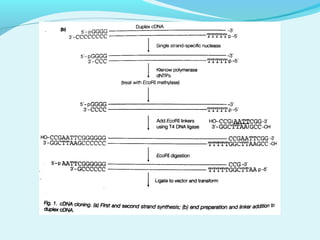
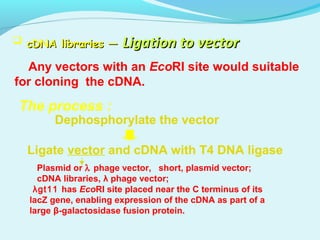
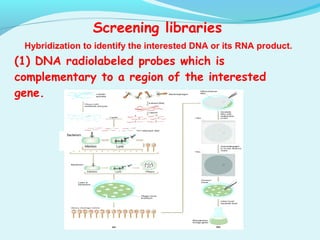
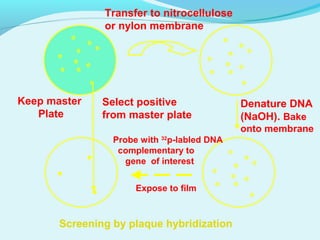
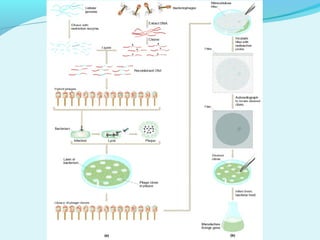
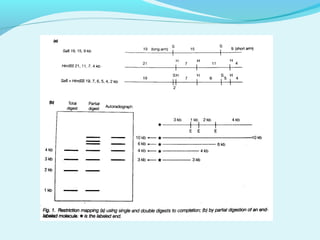
This document discusses genomic and cDNA libraries. Genomic libraries are made from genomic DNA and represent all genes in an organism. They require a minimum number of clones to ensure all genes are captured. cDNA libraries are made from mRNA and represent expressed genes, avoiding introns. Key steps in making cDNA libraries include mRNA isolation, cDNA synthesis, addition of linkers, and ligation into a vector. Screening methods to identify clones of interest include hybridization, expression screening, and hybrid arrest/release.