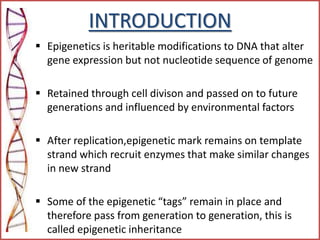
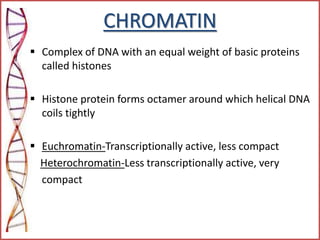
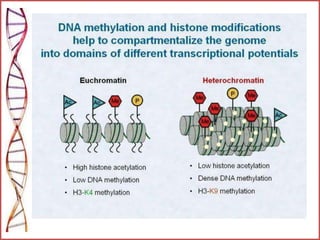
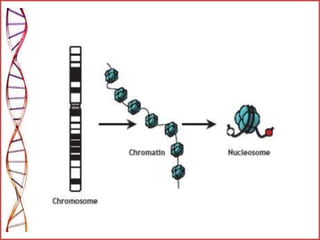
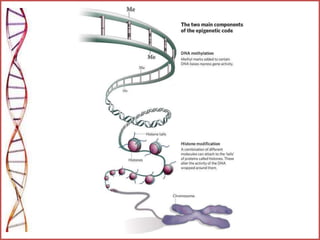
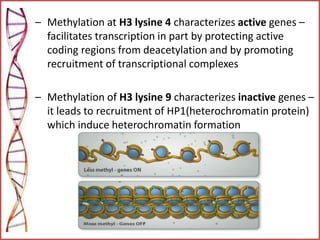
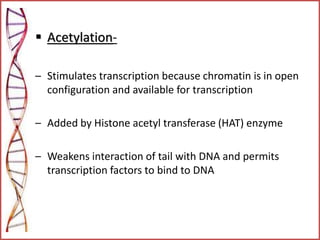
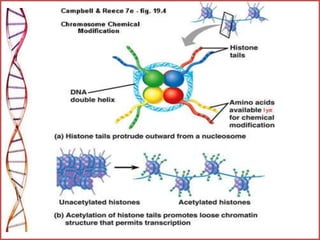
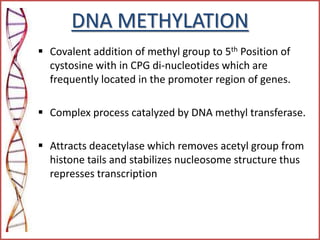
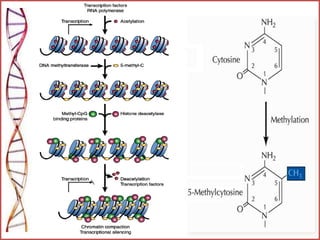
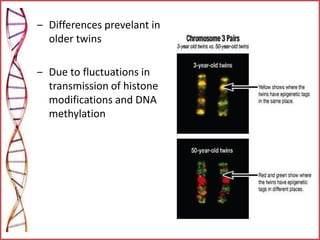
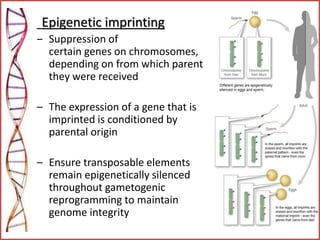
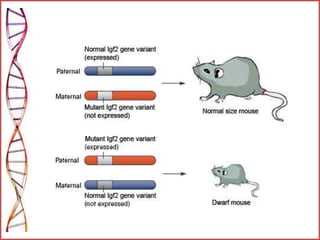
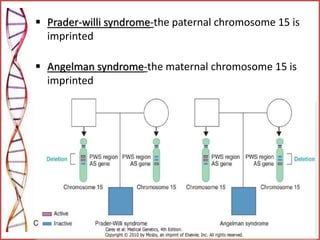
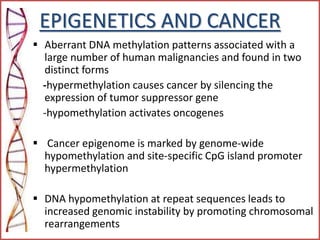
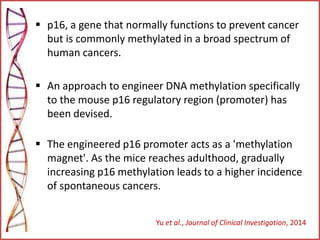
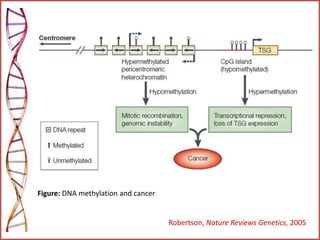
The document provides a comprehensive overview of epigenetics, focusing on heritable DNA modifications that influence gene expression without altering the genetic code, including mechanisms such as DNA methylation and histone modifications. It discusses epigenetic inheritance, differences in identical twins, and the implications of epigenetics for cancer, highlighting how methylation patterns can lead to tumor suppression or oncogene activation. Additionally, it emphasizes the potential of epigenetic pharmaceuticals as alternative treatments for diseases like cancer, showcasing ongoing research in the field.