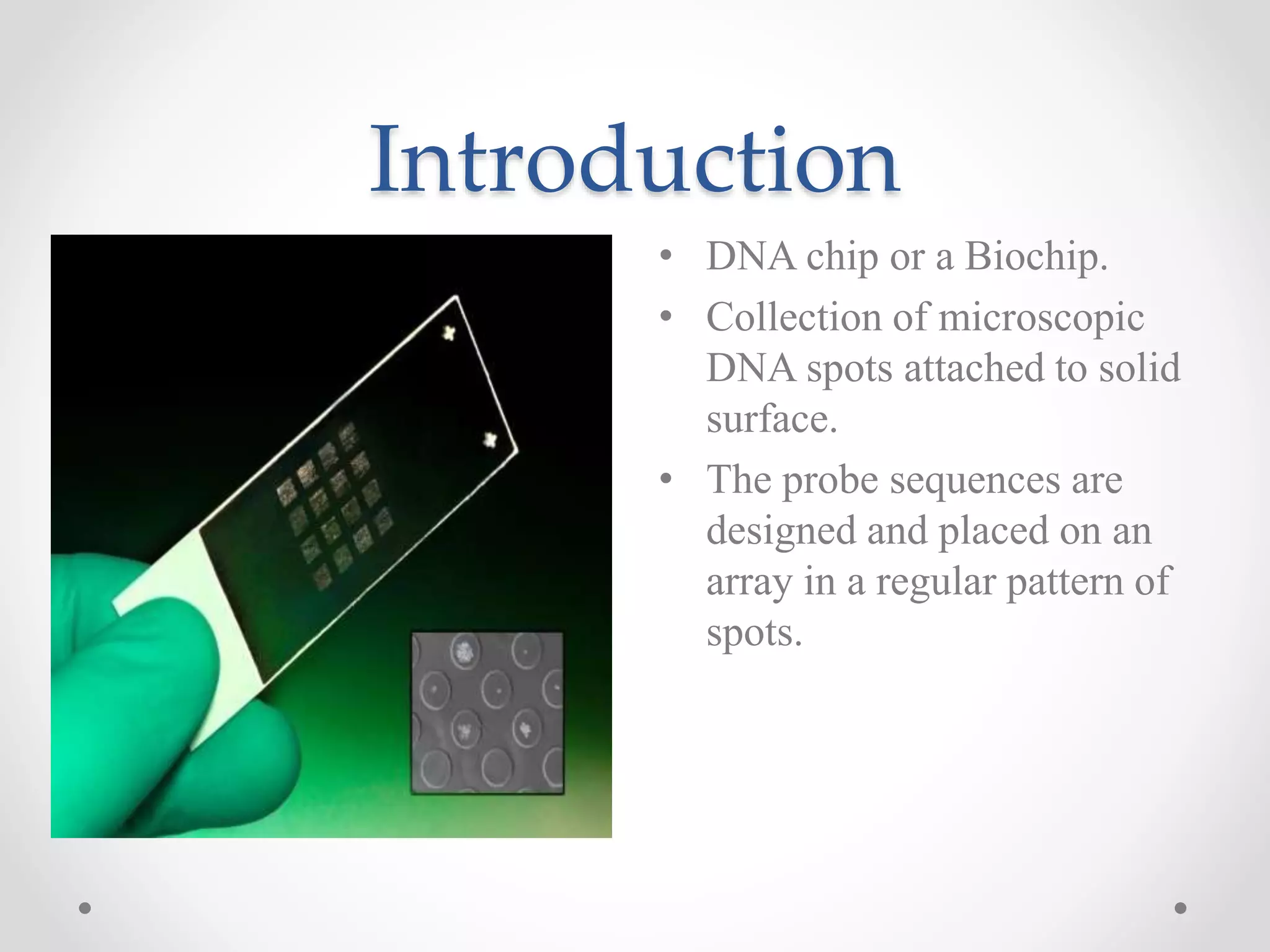
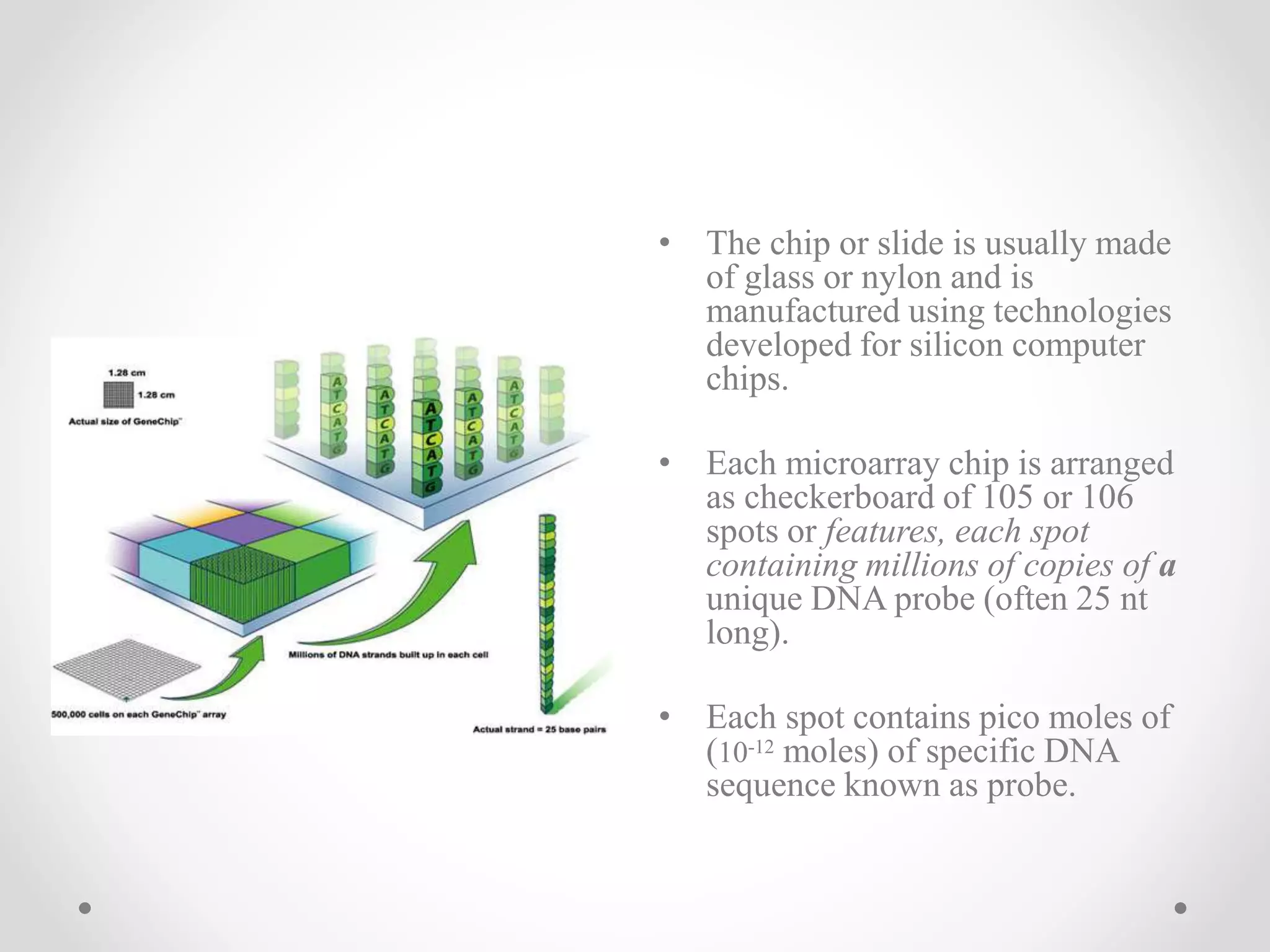
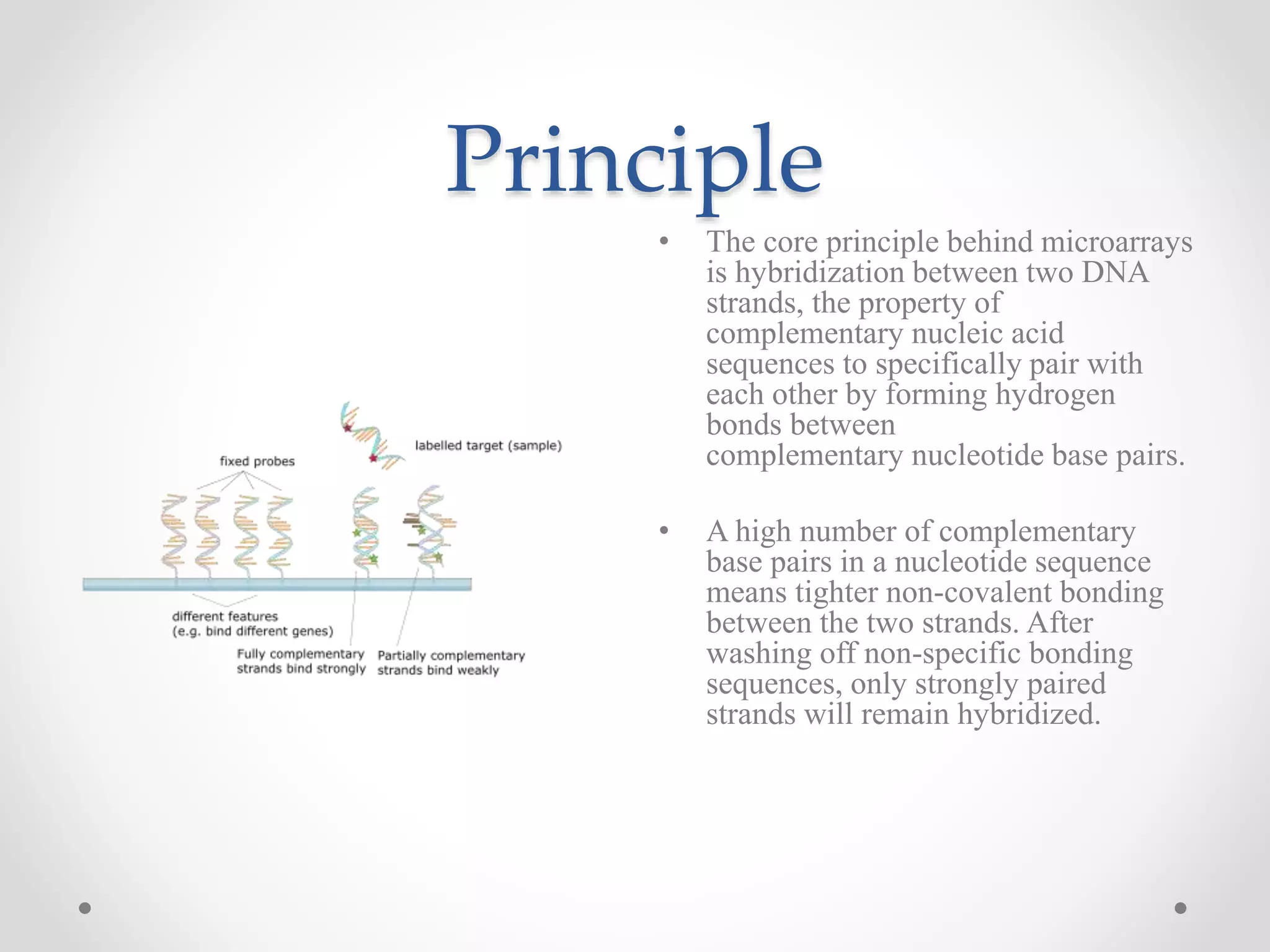
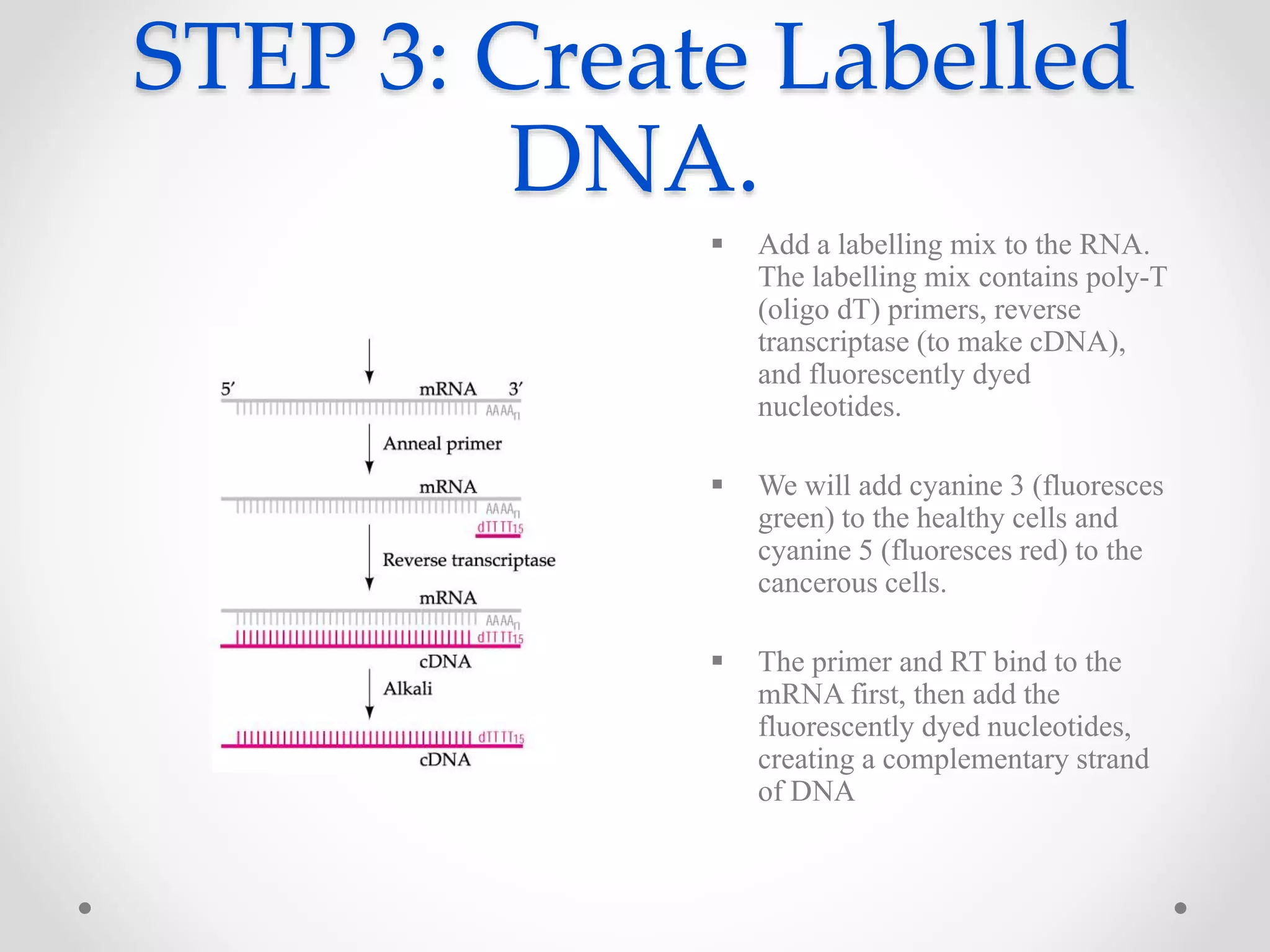
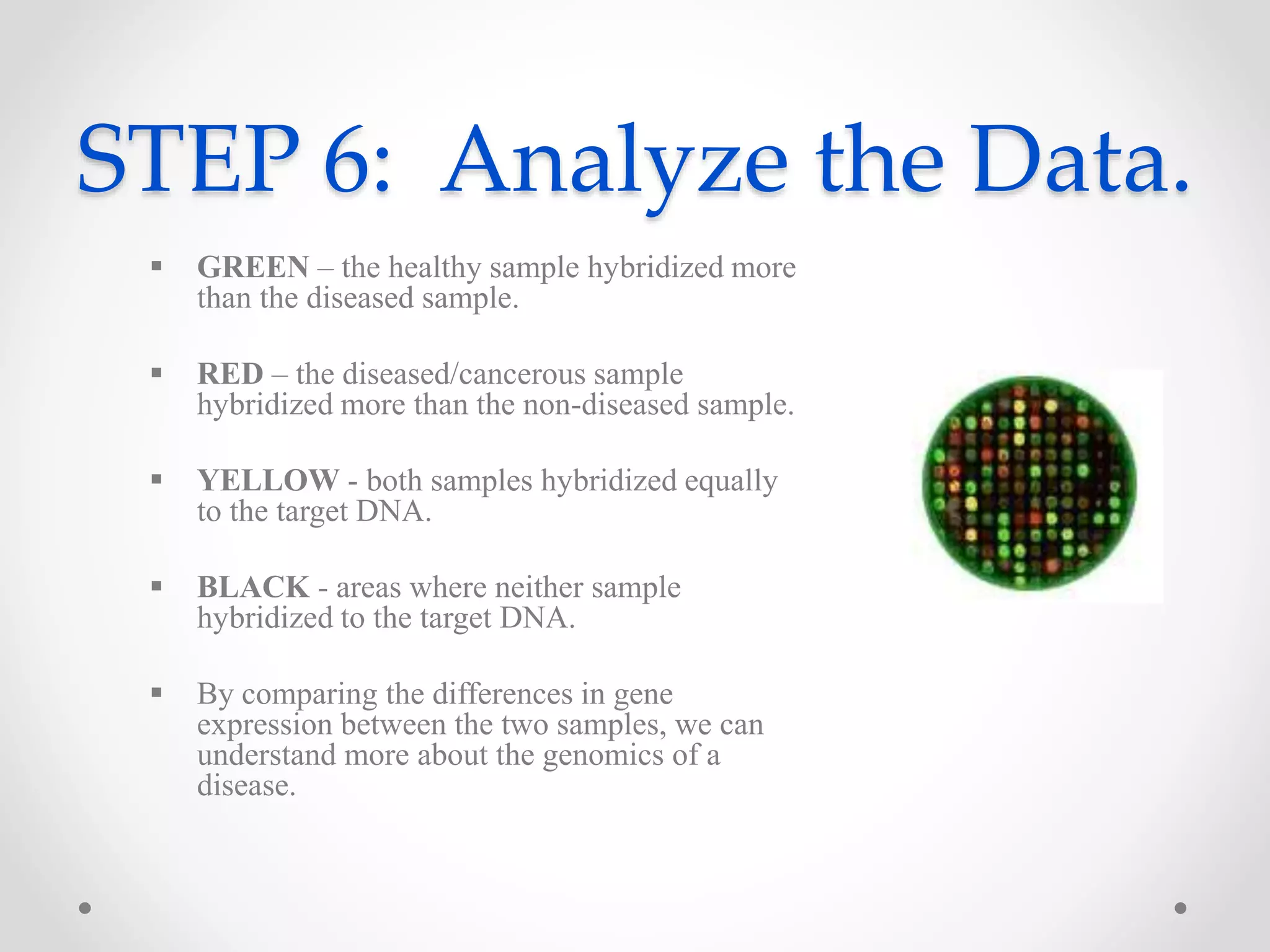
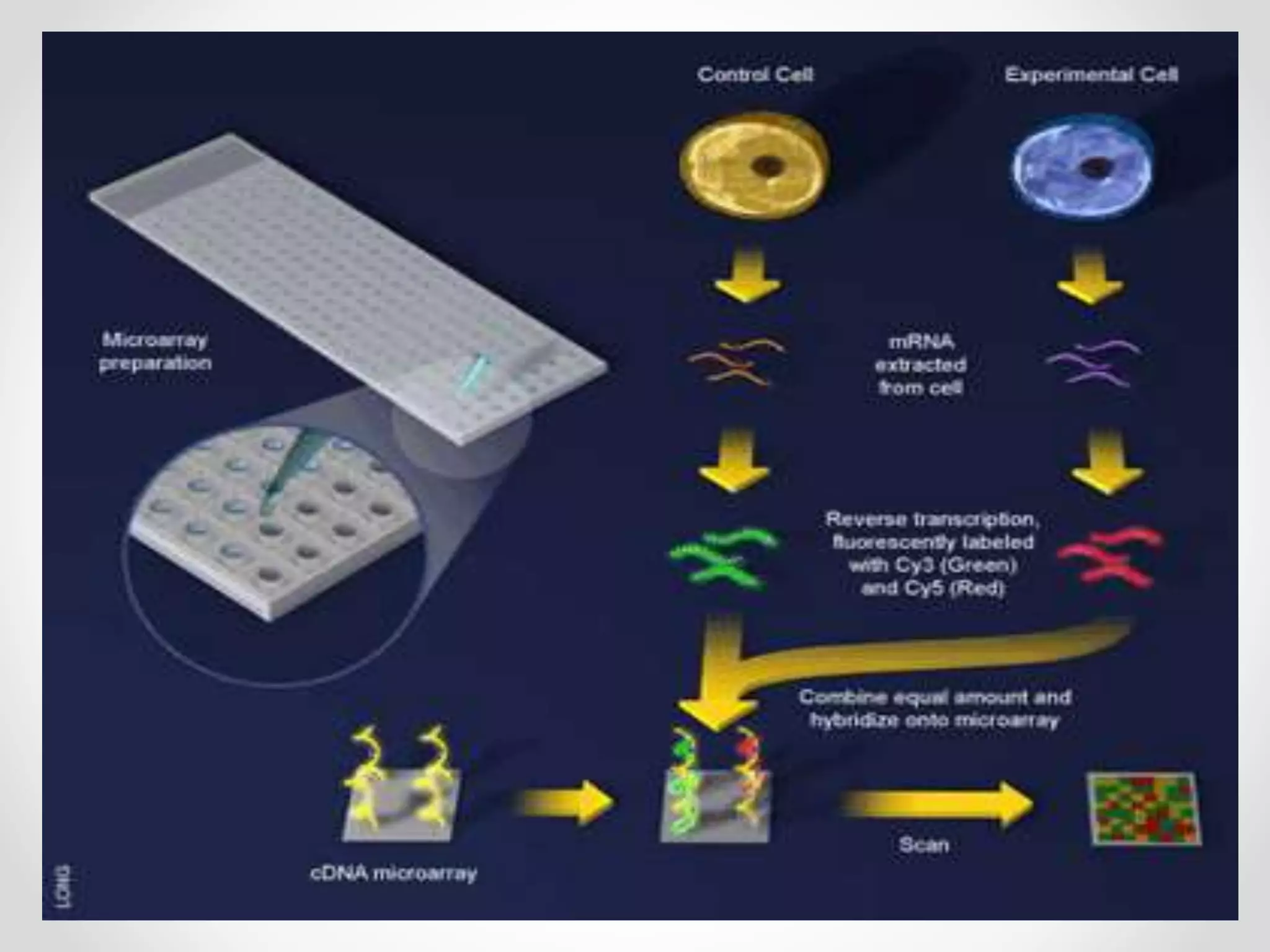
DNA microarrays allow researchers to analyze gene expression levels across thousands of genes simultaneously. A DNA microarray contains many DNA probes attached to a solid surface in a regular pattern. Researchers isolate mRNA from samples, convert it to cDNA, and label the cDNA with fluorescent dyes. They then hybridize the labeled cDNA to the probes on the microarray. A scanner detects the fluorescence at each probe location, allowing researchers to compare gene expression levels between samples by the intensity and color of fluorescence. Microarrays have applications in medicine, agriculture, forensics and toxicology by enabling the comparison of gene expression in different tissues or in response to different conditions.