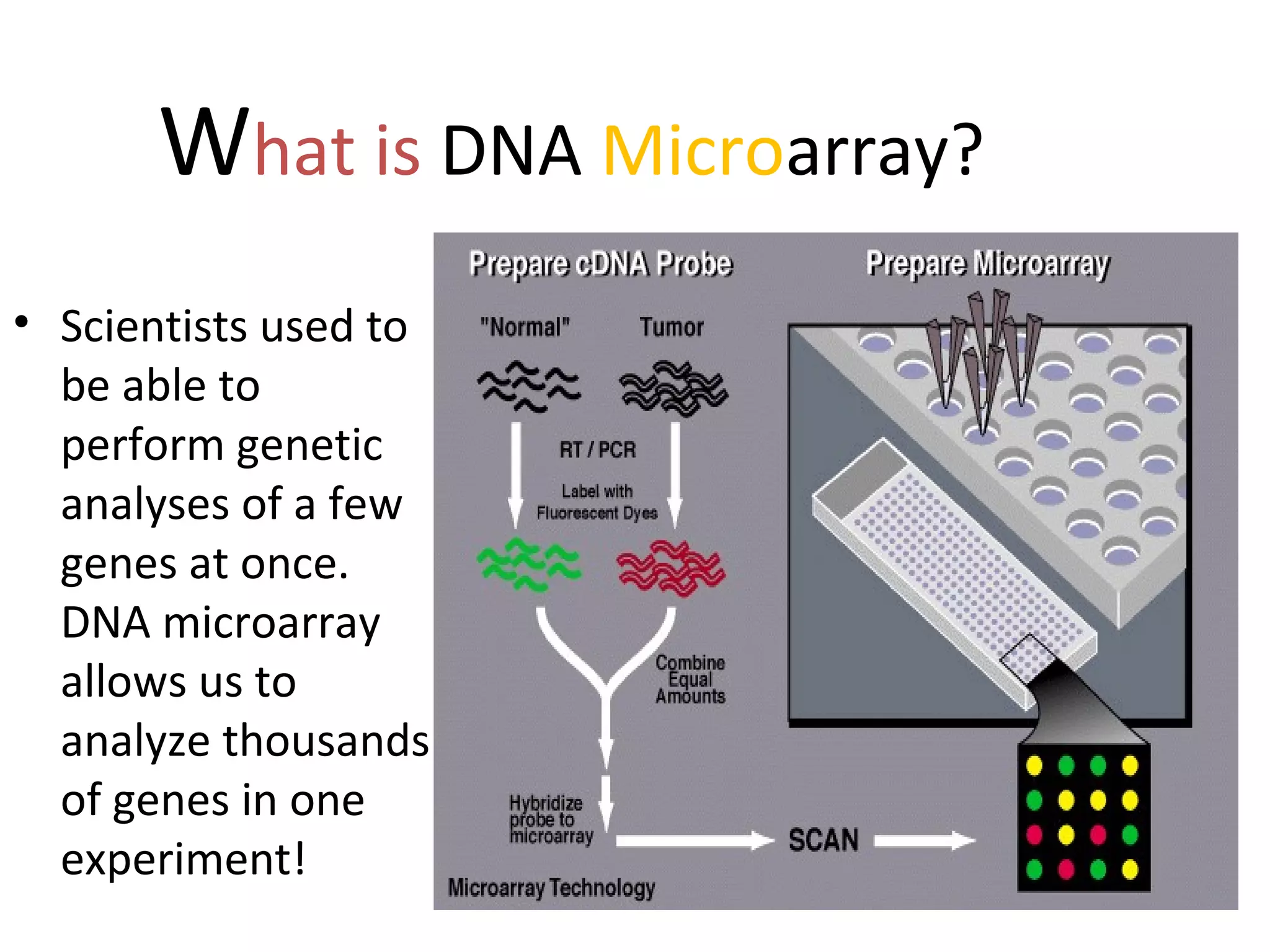
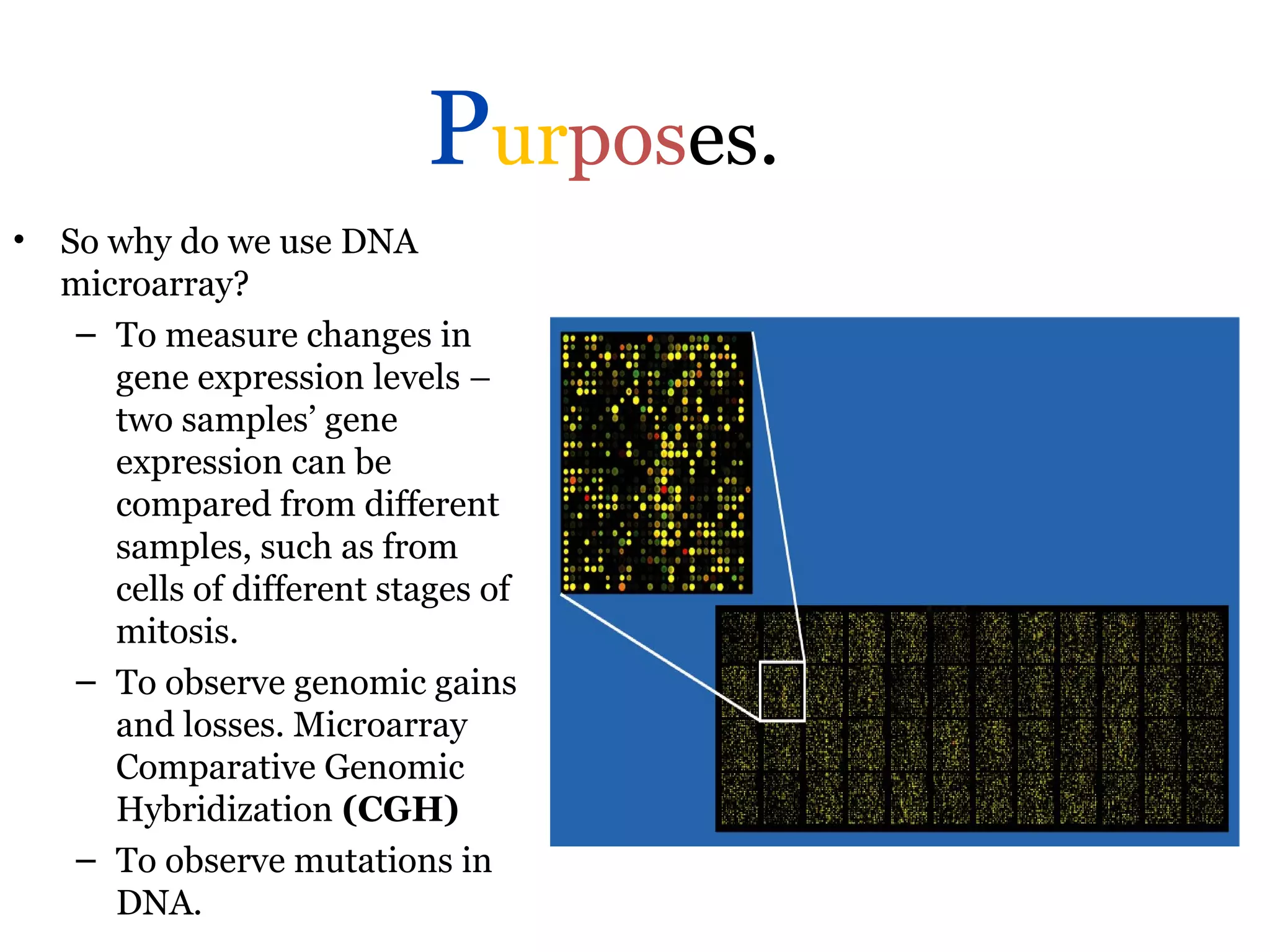
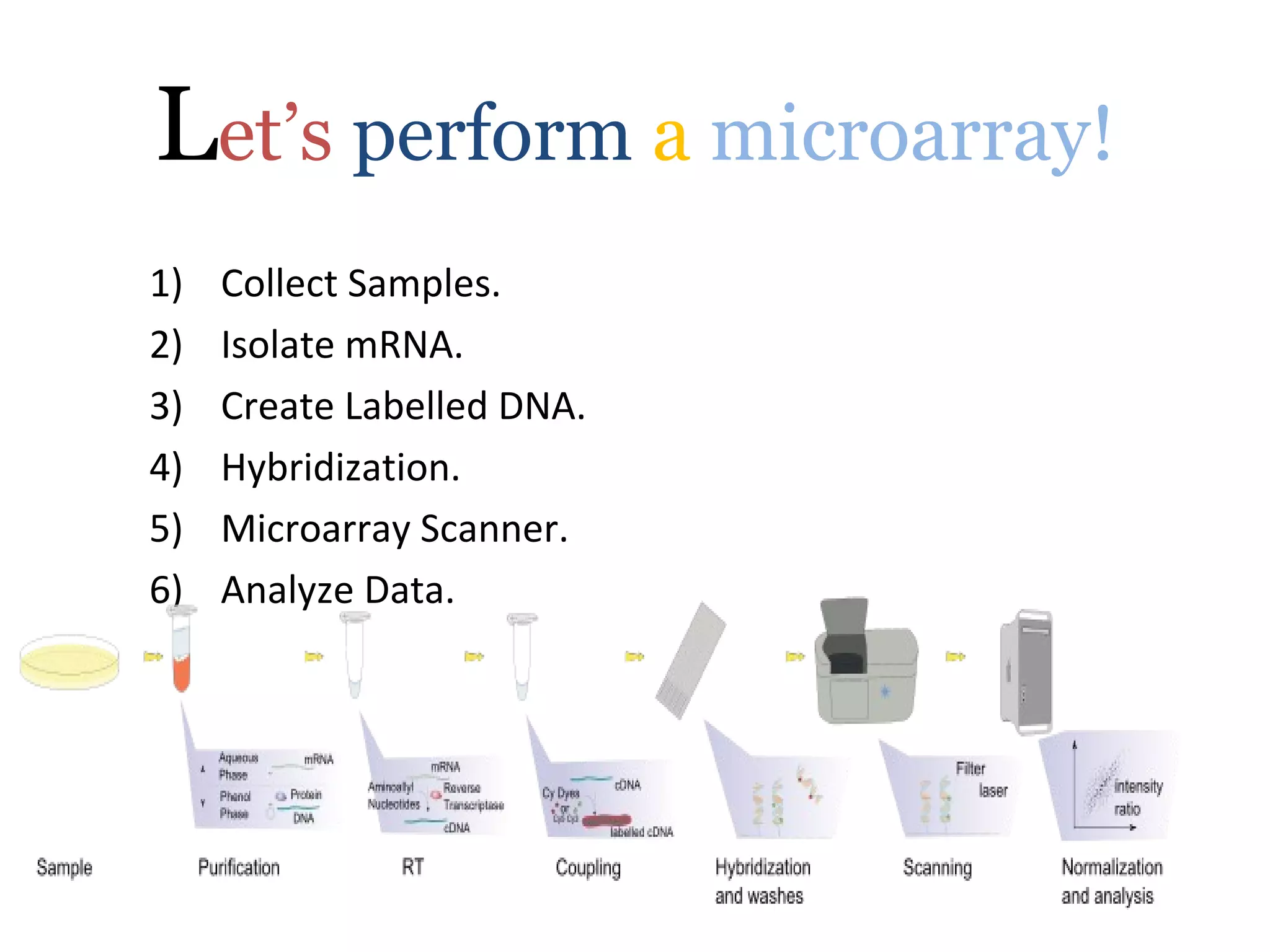
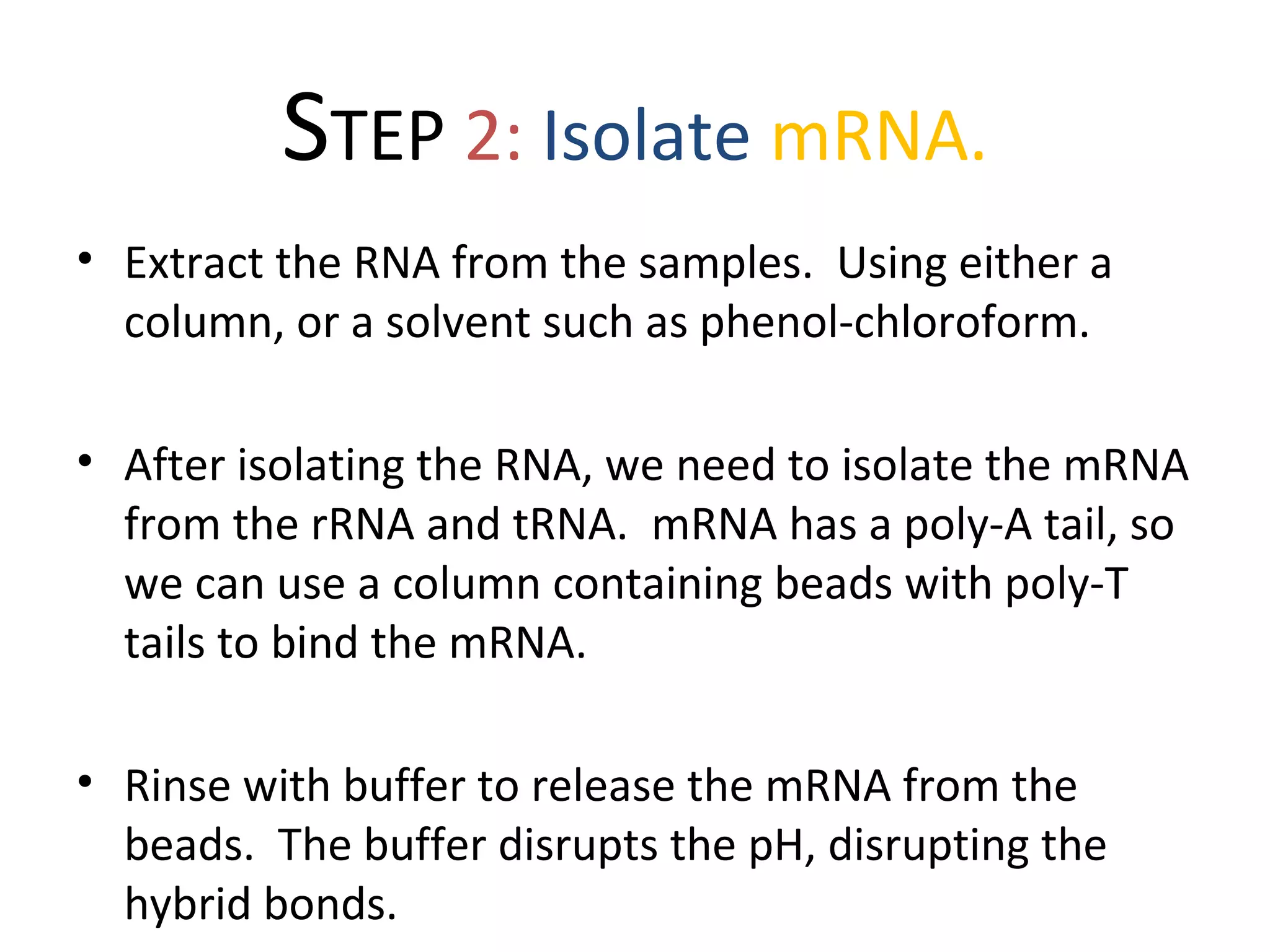
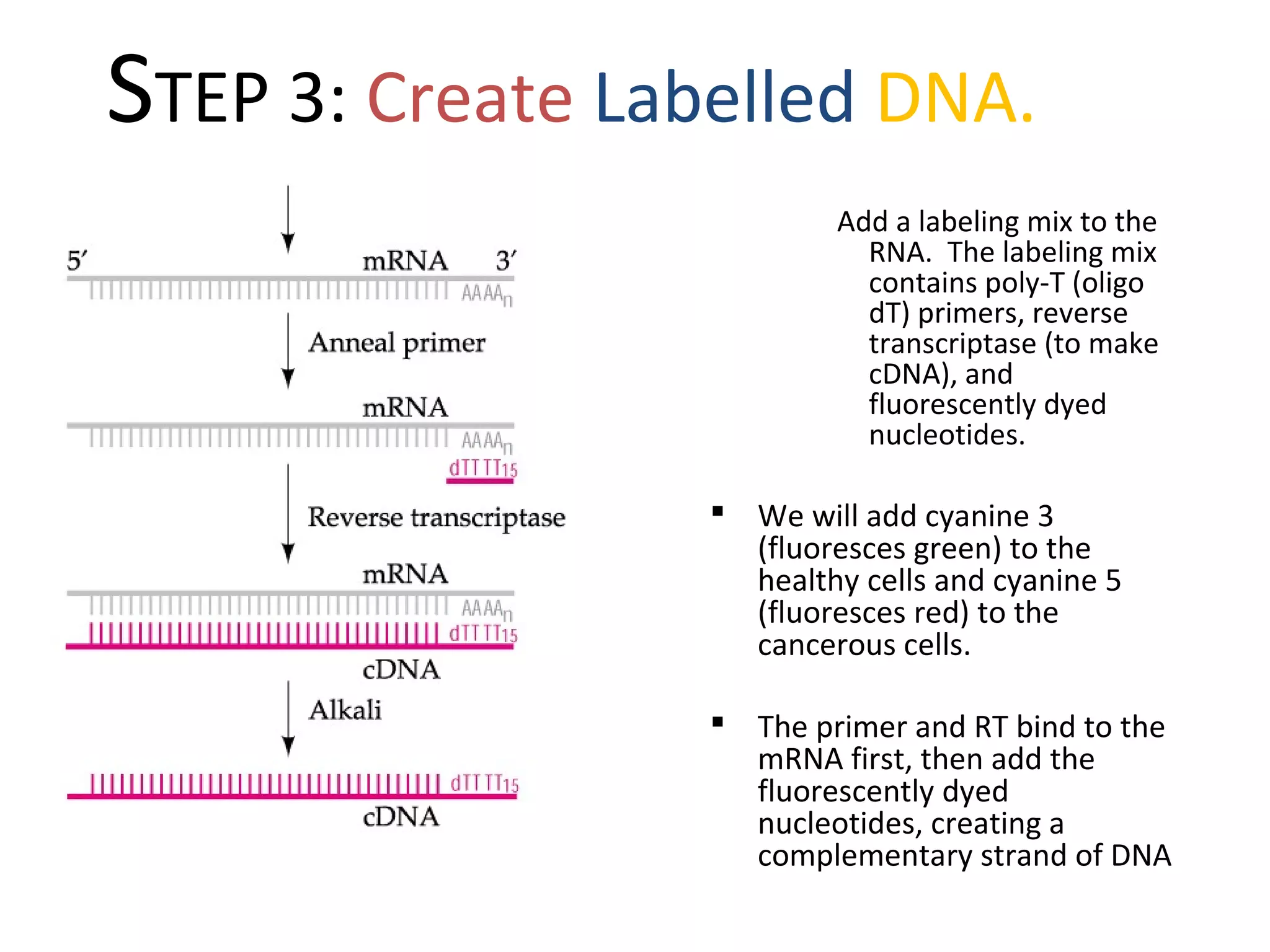
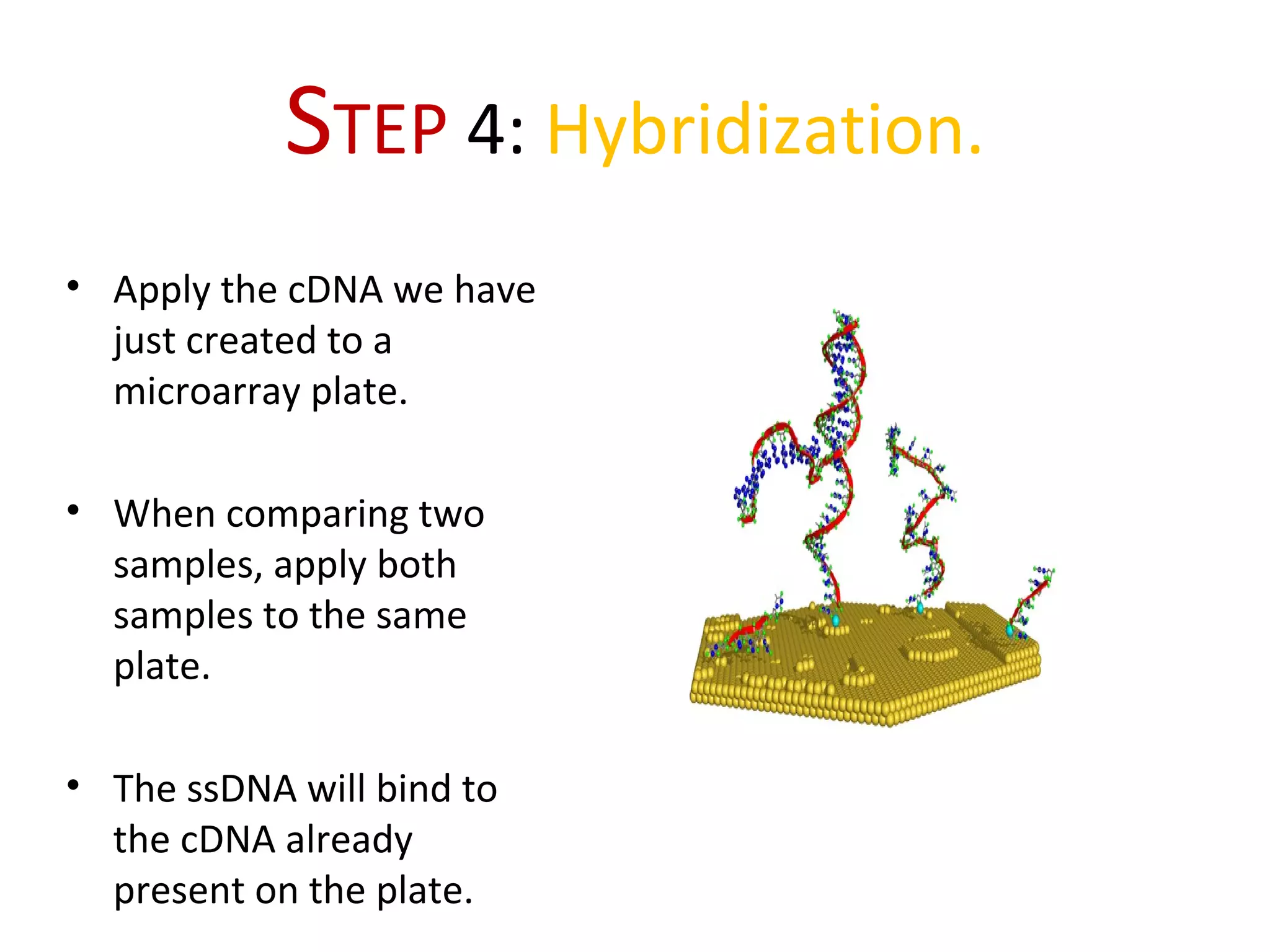
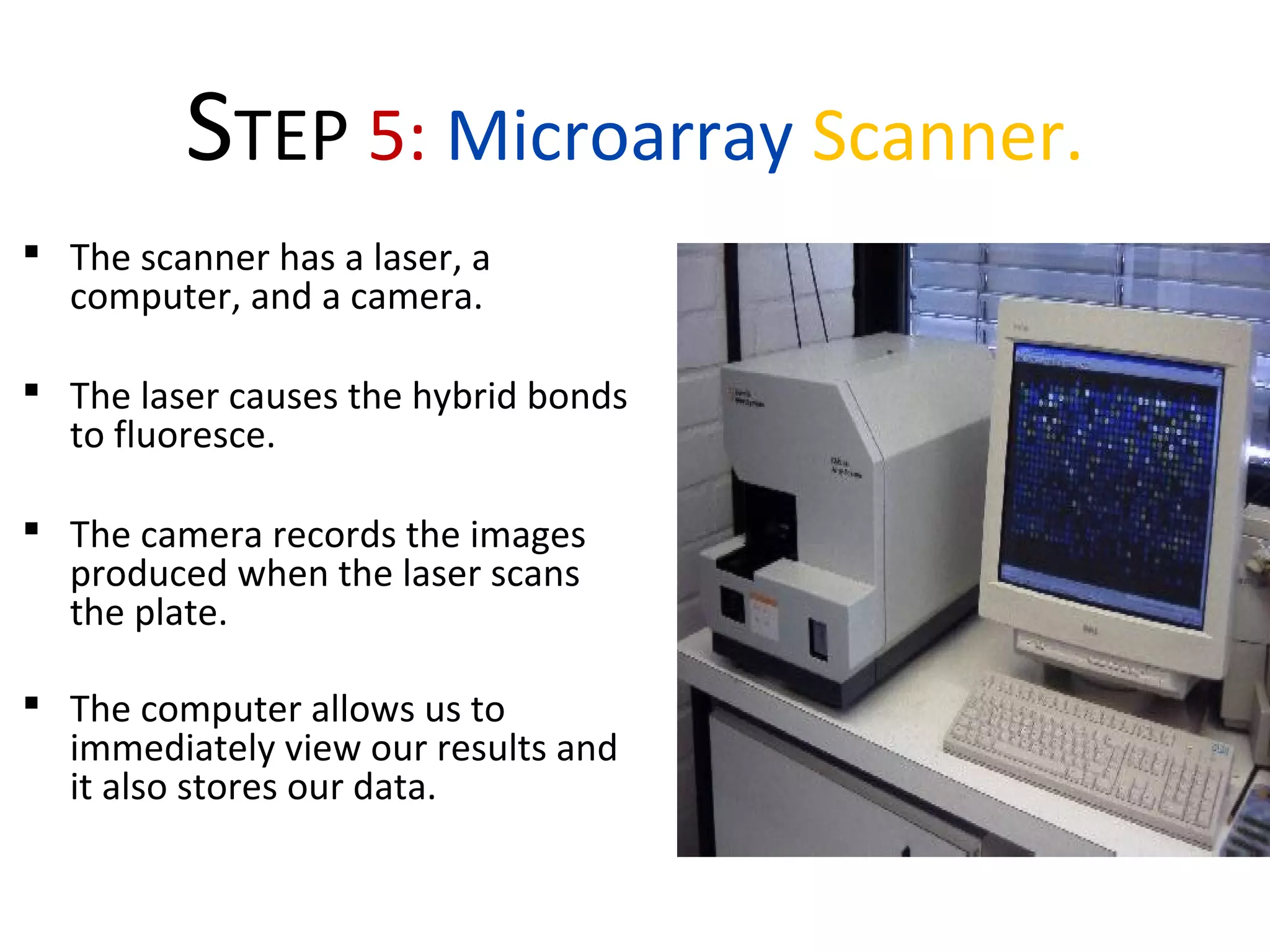
DNA microarrays allow scientists to analyze thousands of genes simultaneously. They work by attaching DNA probes to a plate to measure gene expression levels in different samples. The process involves isolating mRNA from samples, converting it to labeled cDNA, hybridizing it to the microarray plate, and scanning the plate to analyze gene expression differences between samples. Microarrays have benefits like speed and analyzing many genes at once, though they also produce large amounts of data to analyze. Future uses include disease diagnosis, pharmacogenomics, and toxicogenomics research.