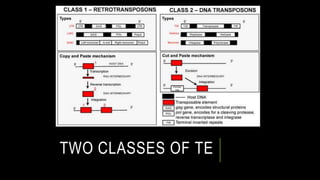
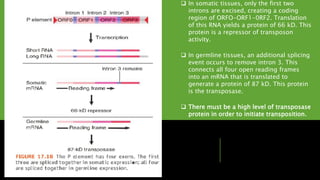
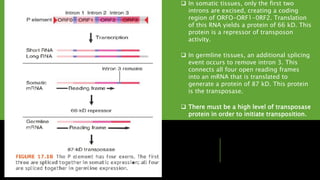
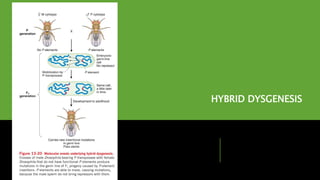
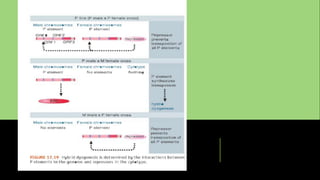
Transposons, or 'jumping genes', are DNA sequences that move within the genome, first discovered by Barbara McClintock in the 1940s. In Drosophila, P elements, a type of Class II transposable elements, can induce hybrid dysgenesis when crossed with M strain flies, leading to genetic defects and reduced fertility. The mechanism of transposition involves a protein called transposase, which is crucial for the activity of P elements, particularly in germline cells where specific splicing events occur.