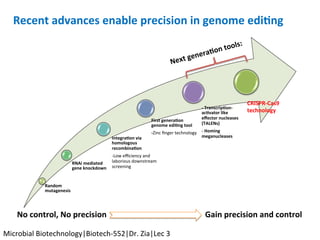
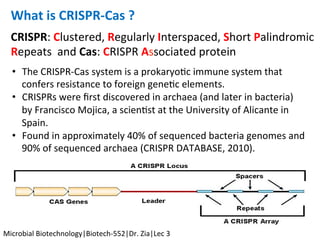
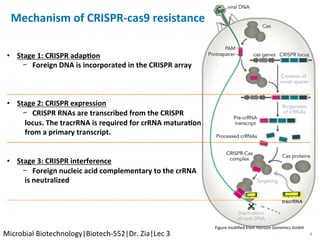
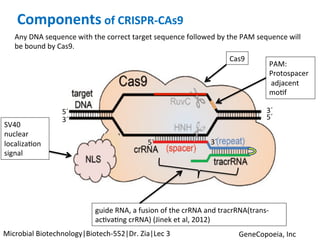
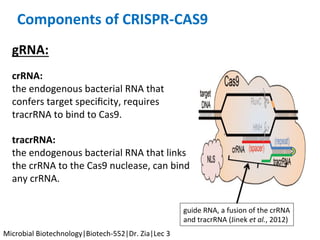
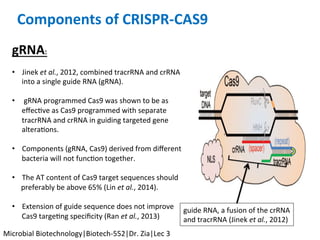
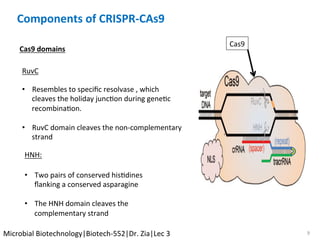
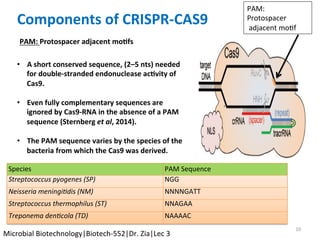
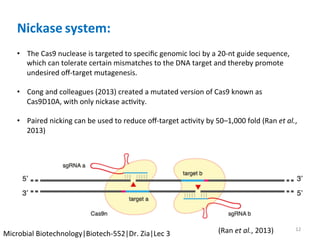
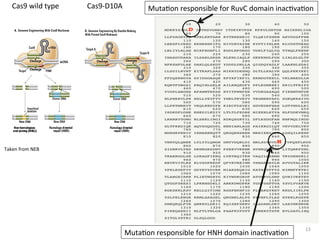
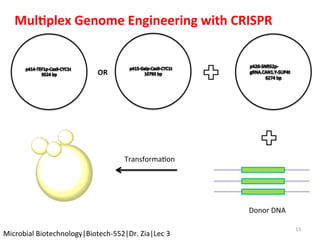
The document discusses the CRISPR-Cas9 system and its use as a genome editing tool. It provides background on the discovery of CRISPRs in bacteria and archaea. It describes the mechanism of CRISPR-Cas9 resistance, including adaptation, expression and interference stages. It discusses the components of CRISPR-Cas9 including the guide RNA and Cas9 nuclease domains. It also covers the protospacer adjacent motif (PAM) sequence, DNA repair pathways used, applications of nickase systems and multiplex genome engineering with CRISPR.