Embed presentation
Download to read offline


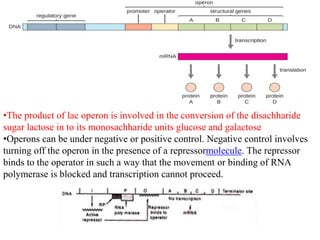
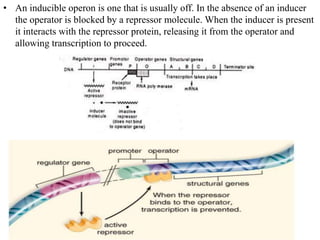

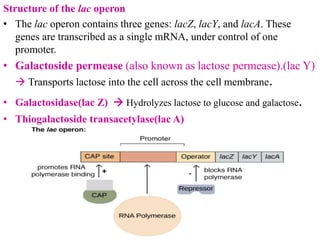
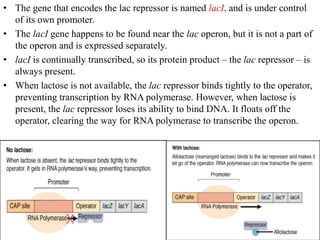
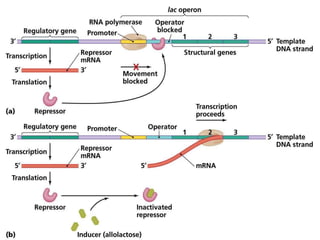
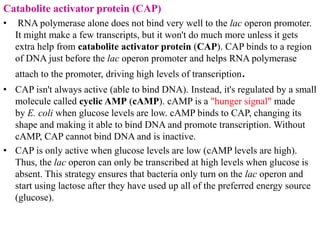

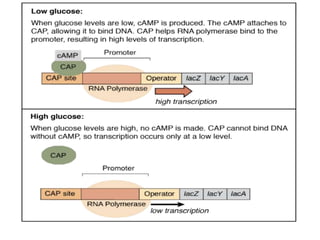

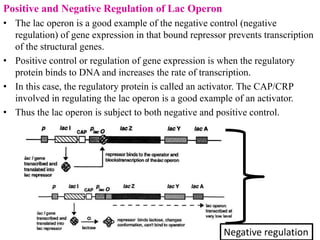
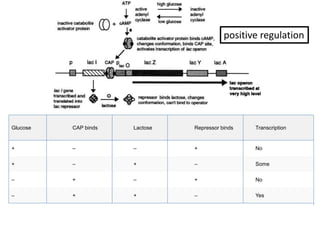
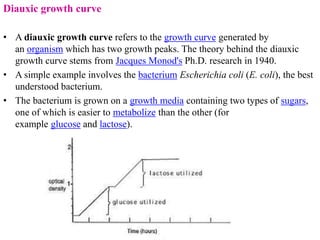


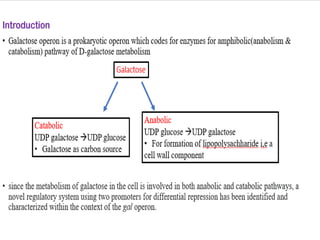
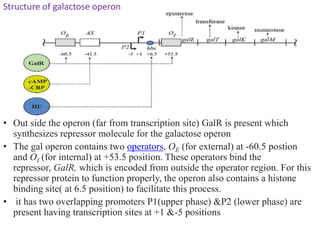

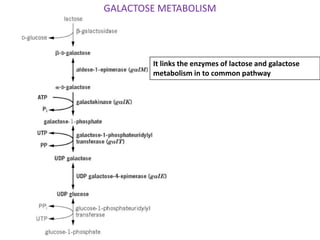
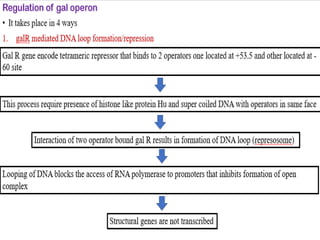
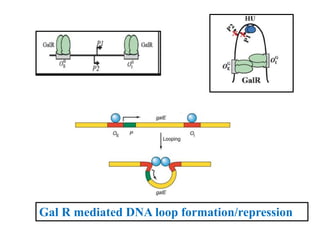
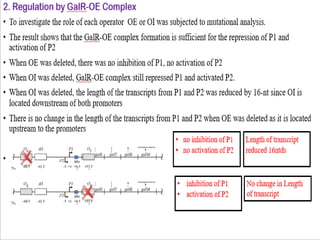
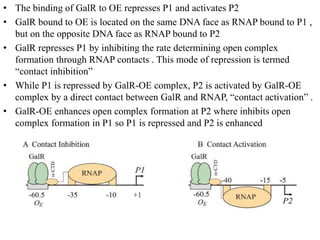
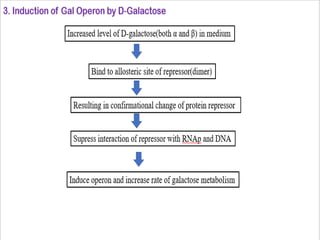
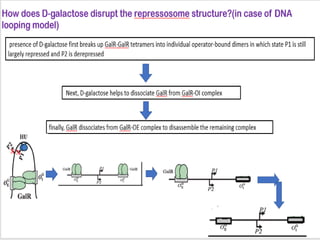
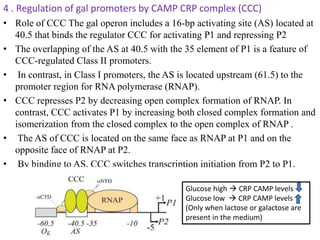


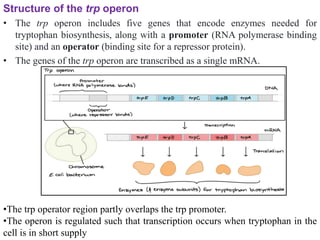
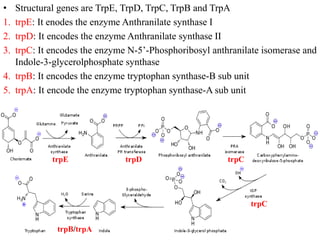
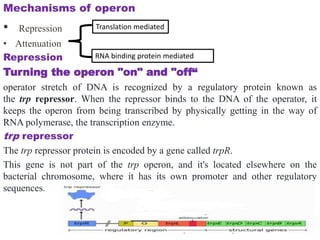
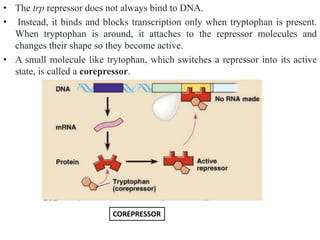
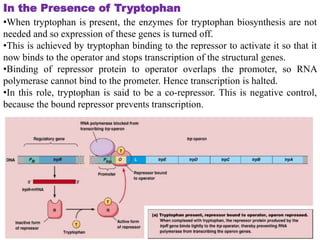
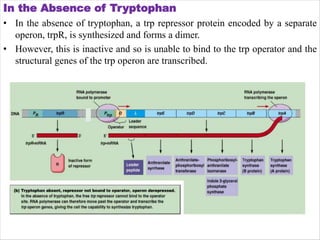
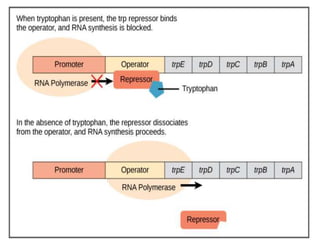
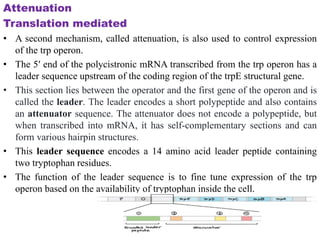
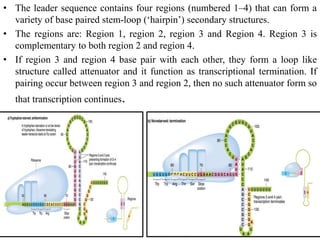
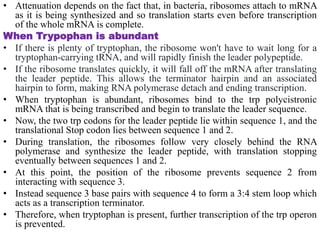
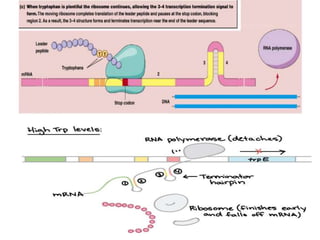

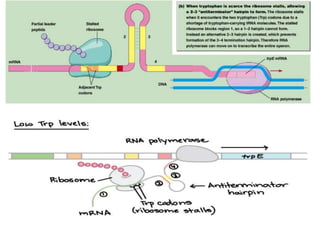
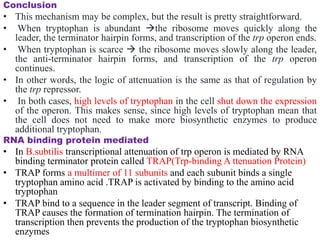


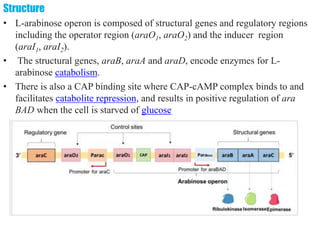
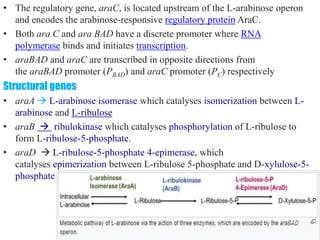
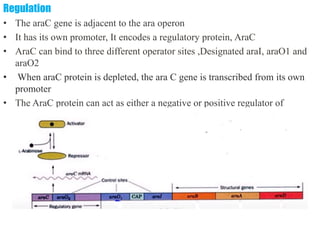
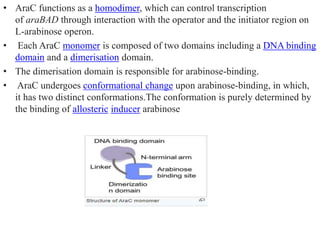
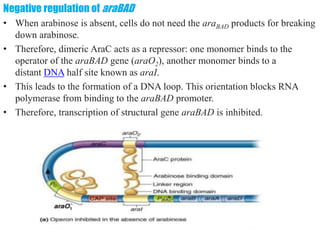
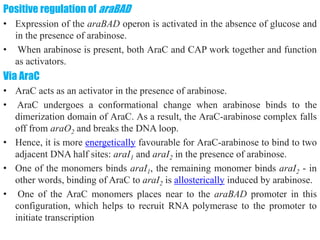
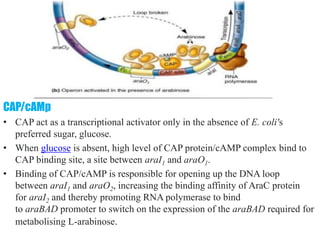
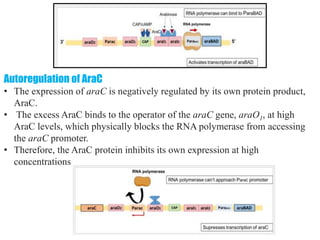


The document discusses the operon model, which explains gene regulation at the transcription level in prokaryotes, specifically focusing on the lac and galactose operons. It details the components and functions of these operons, including how they respond to the presence of sugars like glucose and lactose and the mechanisms of negative and positive control of transcription. Additionally, it describes the trp operon, which is involved in tryptophan biosynthesis, highlighting how its expression is regulated based on the availability of tryptophan in the environment.




























































