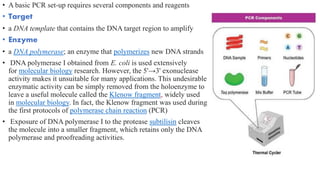
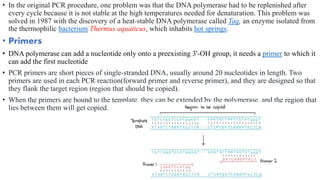
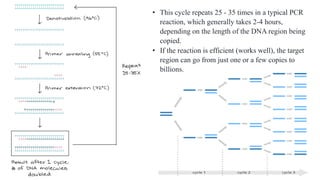
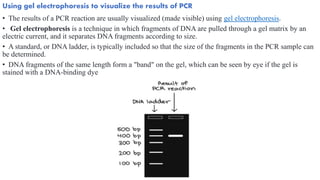
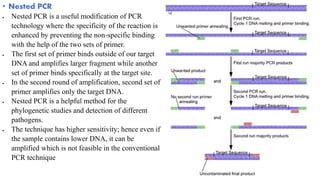
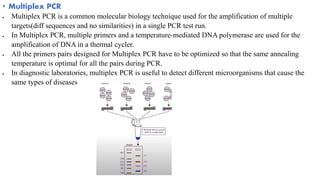
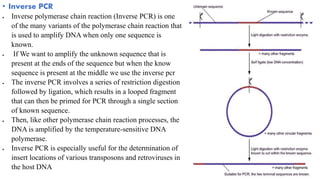
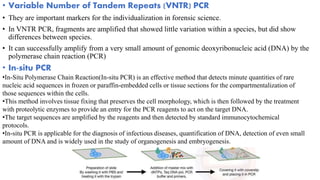
The document provides an overview of Polymerase Chain Reaction (PCR), detailing its invention by Kary Mullis in 1983, the basic principles of DNA amplification, and the necessary components for PCR setup. It describes the cyclic process of denaturation, annealing, and elongation, as well as various applications of PCR in fields such as genetics, forensics, and diagnostics. Additionally, the text explores several PCR variations, including hot start, nested, multiplex, and reverse transcriptase PCR, each with specific uses and benefits.