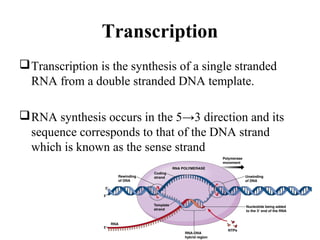
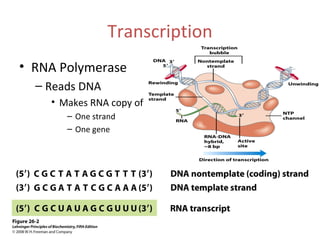
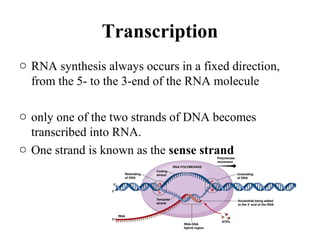
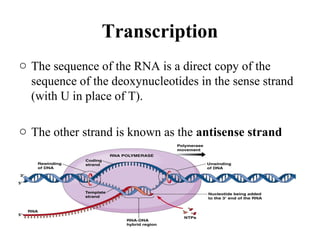
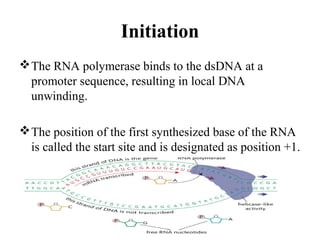
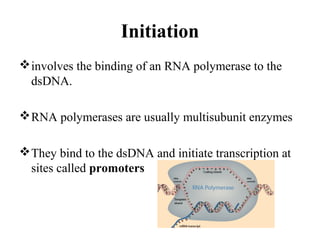
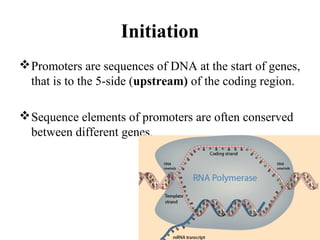
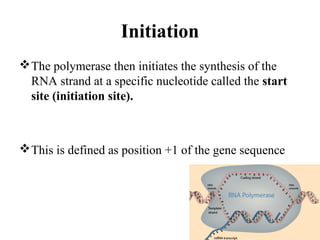
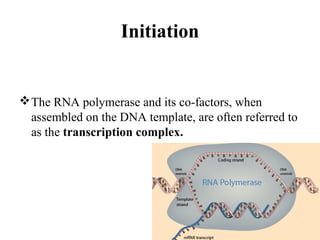
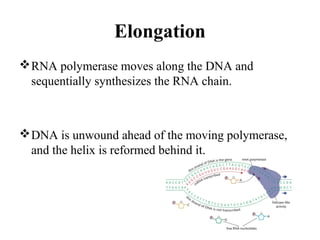
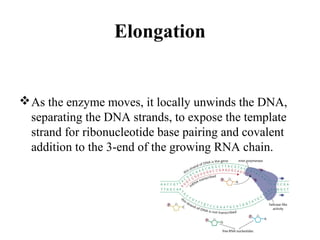
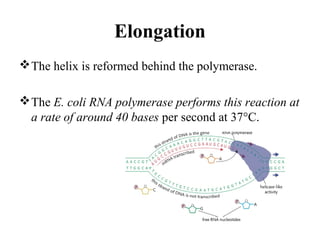
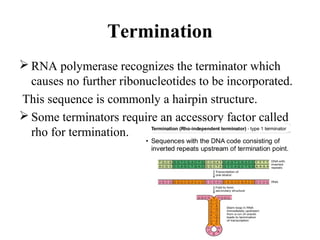
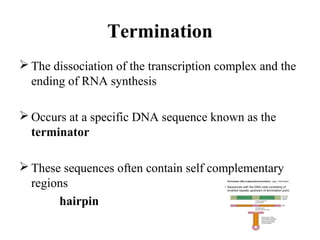
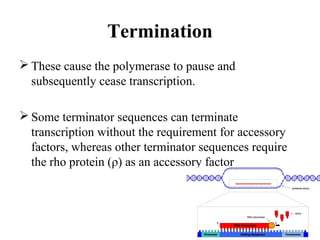
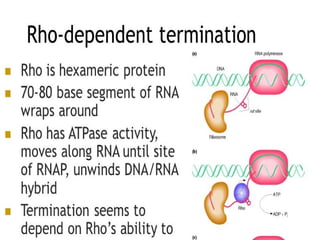
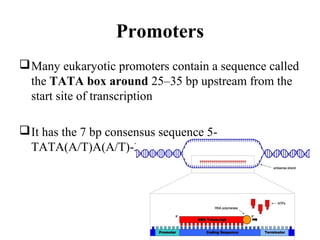
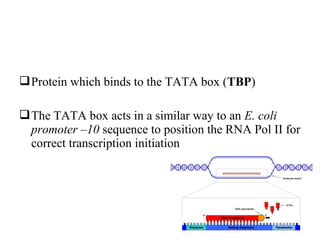
Transcription is the process of synthesizing RNA using a DNA template. It involves three main steps - initiation, elongation, and termination. In initiation, RNA polymerase binds to promoter sequences on DNA and unwinds the double helix. In elongation, RNA polymerase moves along the DNA strand and adds complementary RNA nucleotides. Termination occurs when the polymerase reaches a terminator sequence and stops adding nucleotides. Prokaryotes and eukaryotes have similar transcription mechanisms but eukaryotes have three RNA polymerases and require additional transcription factors.