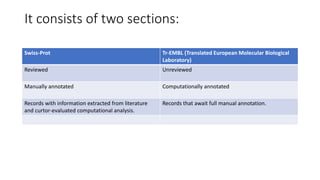
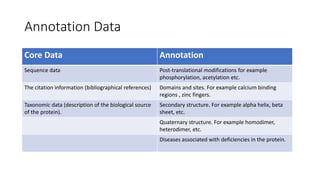
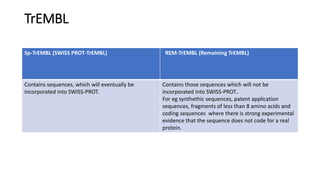
Swiss-Prot is a central hub within the UniProtKB knowledgebase that provides curated functional information on proteins, established as a collaboration between the University of Geneva and the European Molecular Biology Laboratory since 1987. It includes manually reviewed records in Swiss-Prot and unreviewed, computer-annotated records in TrEMBL, allowing for minimal redundancy and integration with other databases. The continuous enhancement of Swiss-Prot's content and format reflects its commitment to high-quality annotation and user accessibility in proteomics.