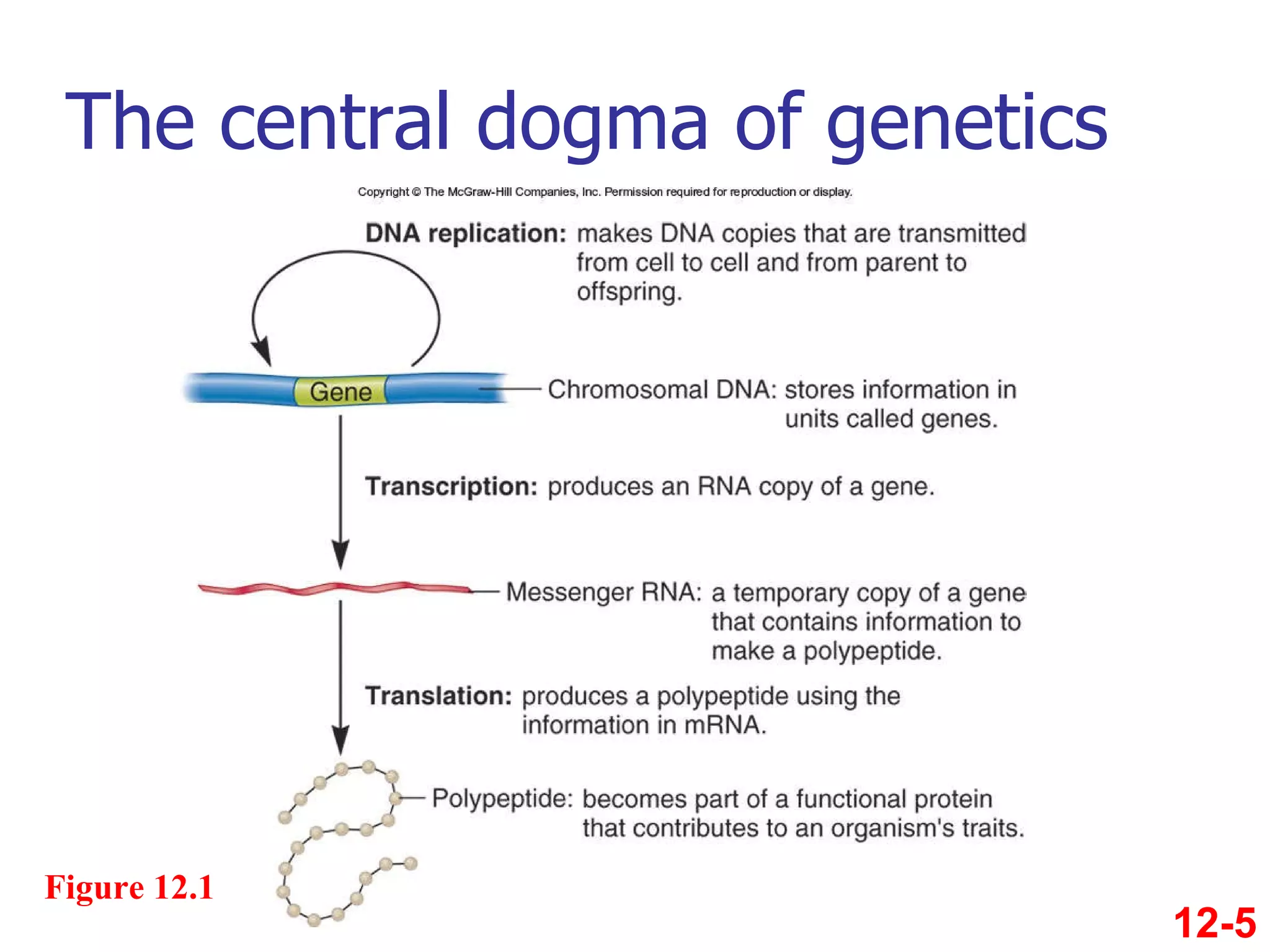
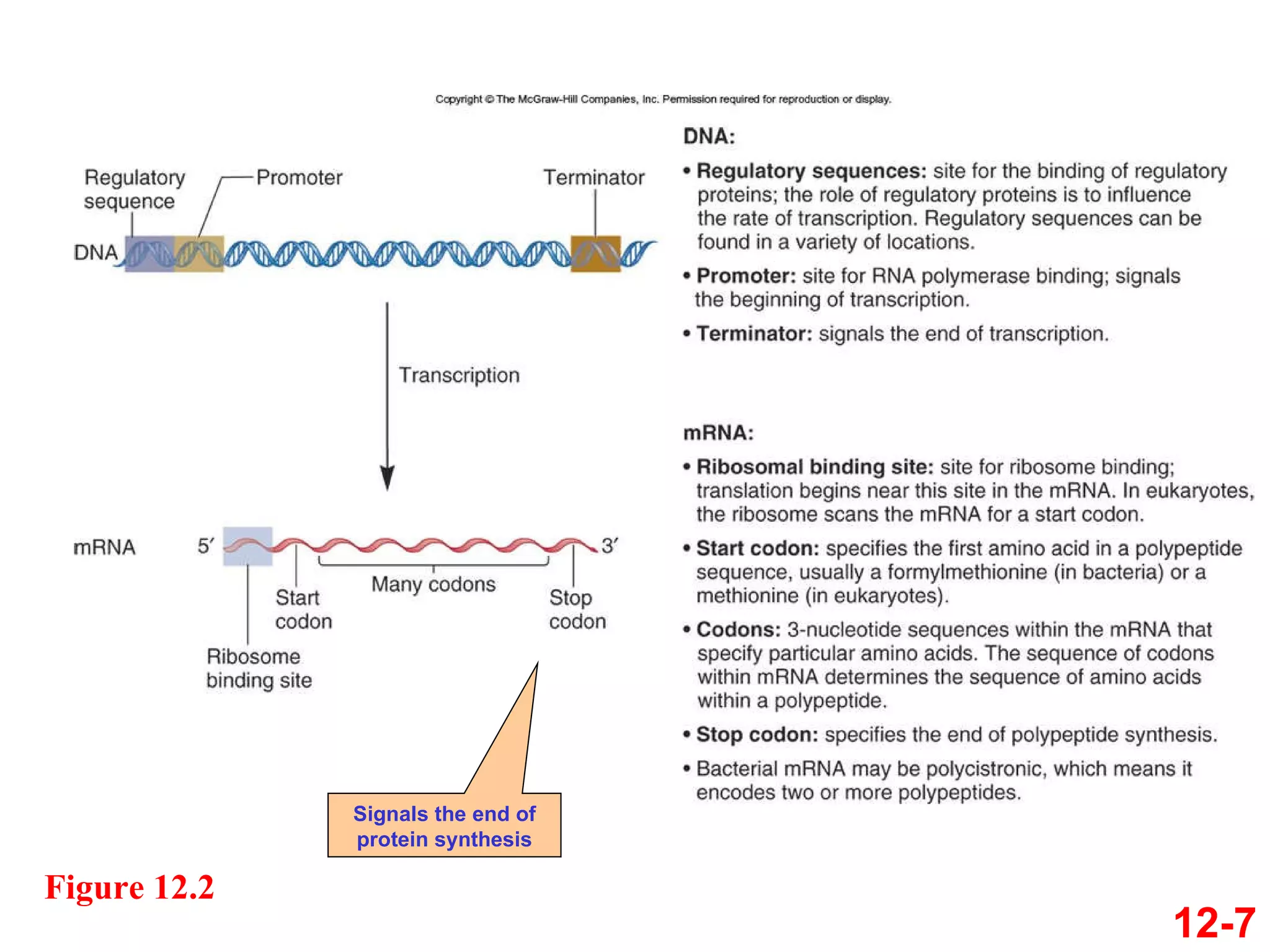
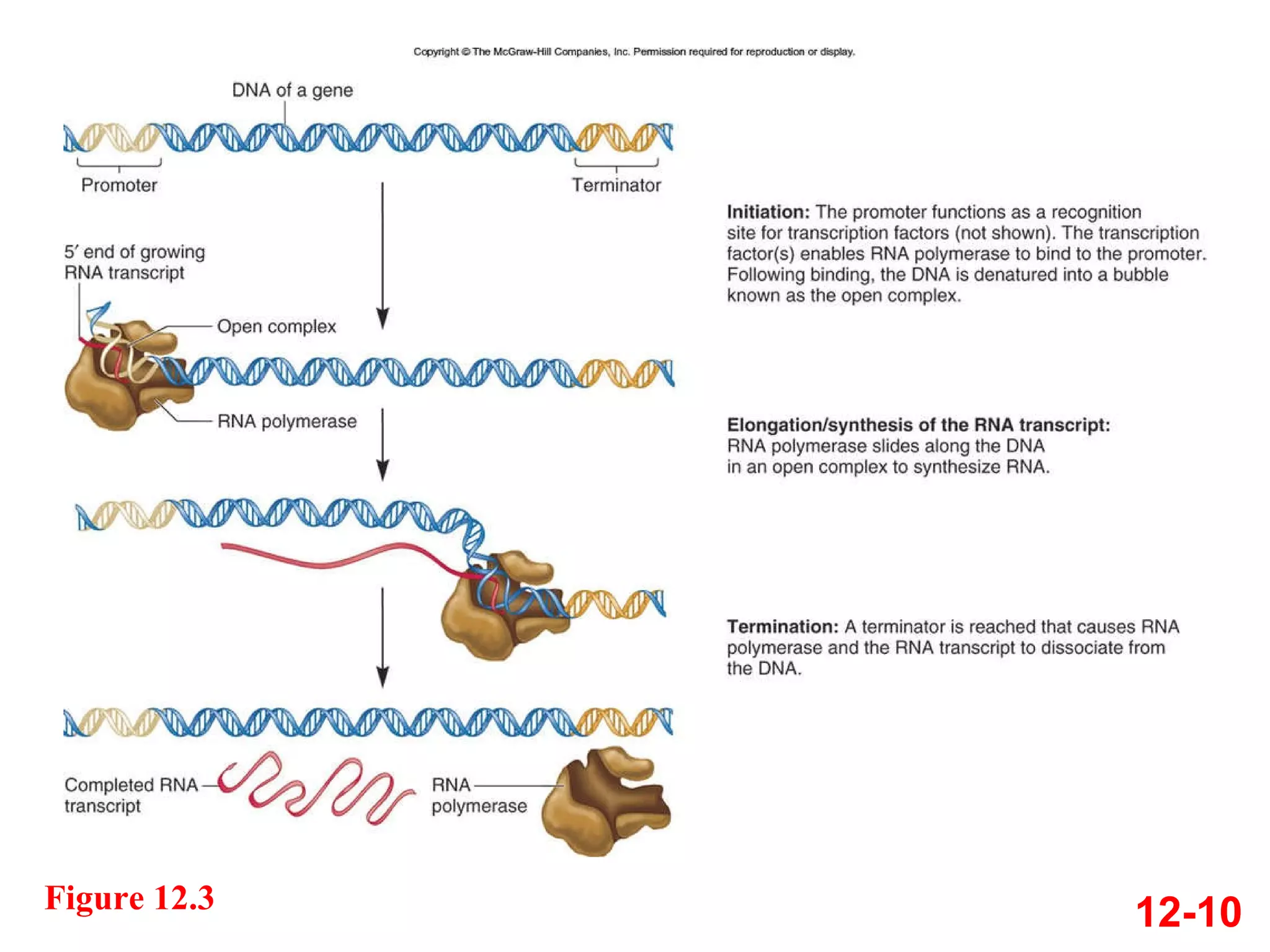
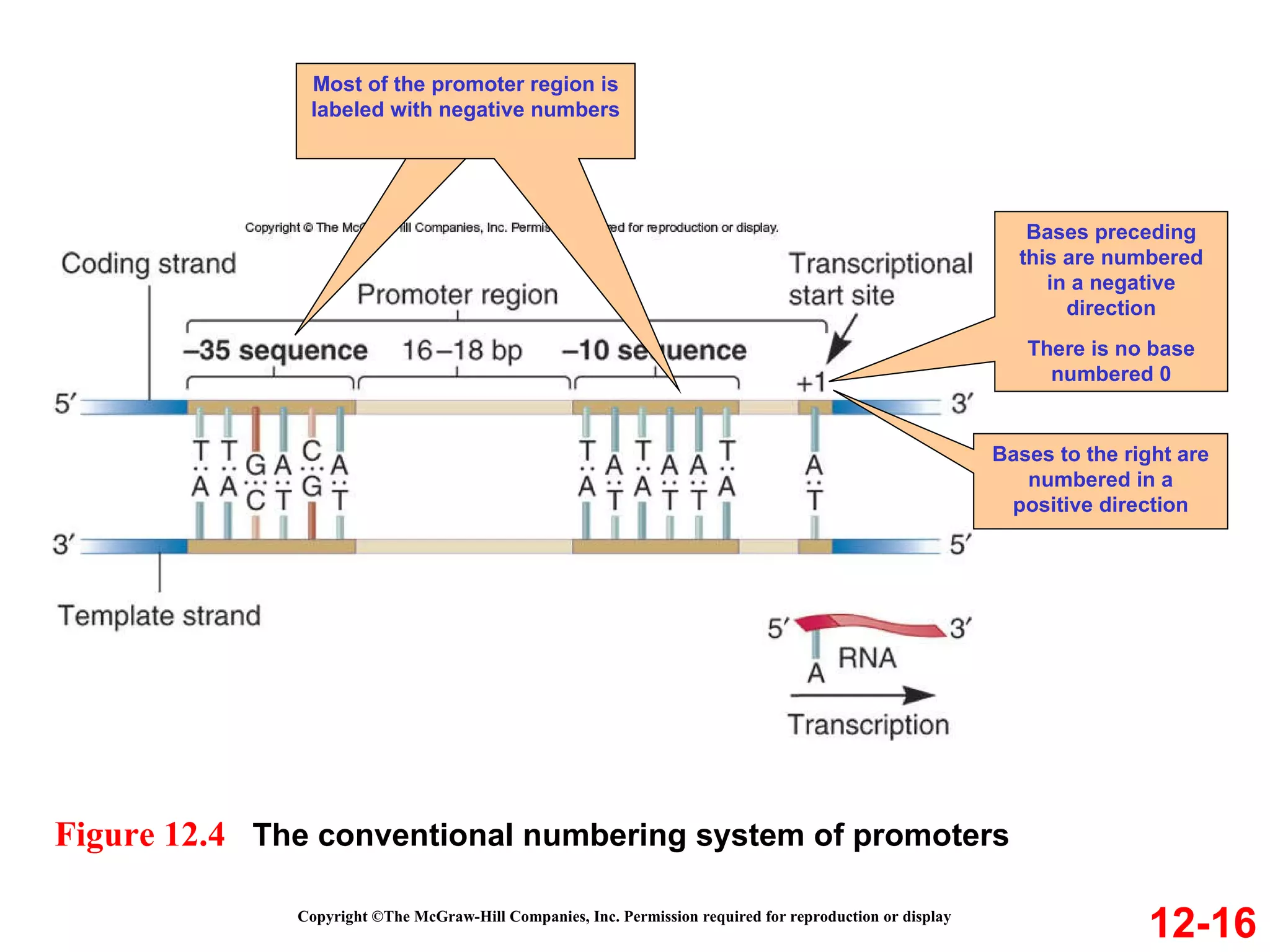
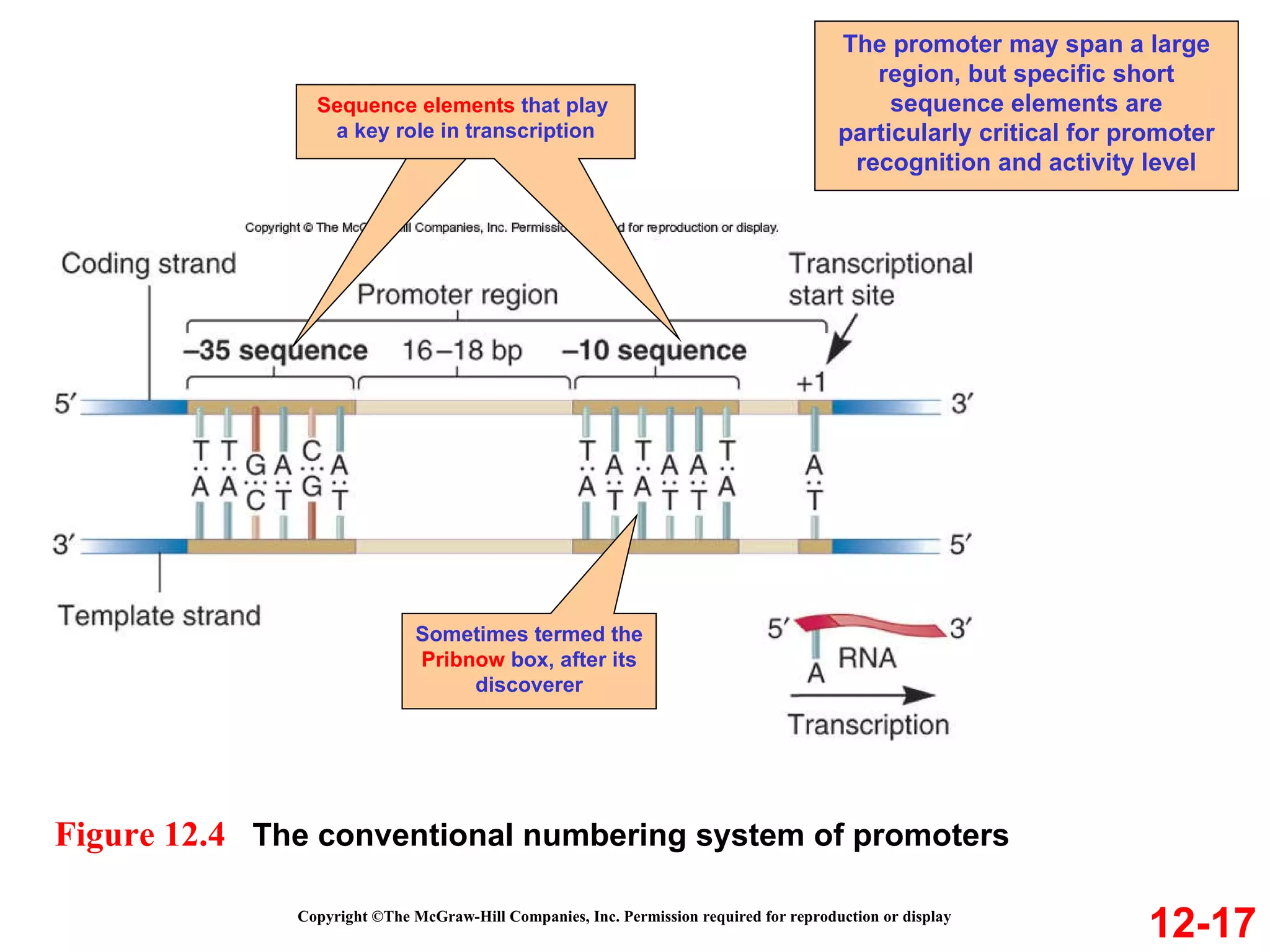
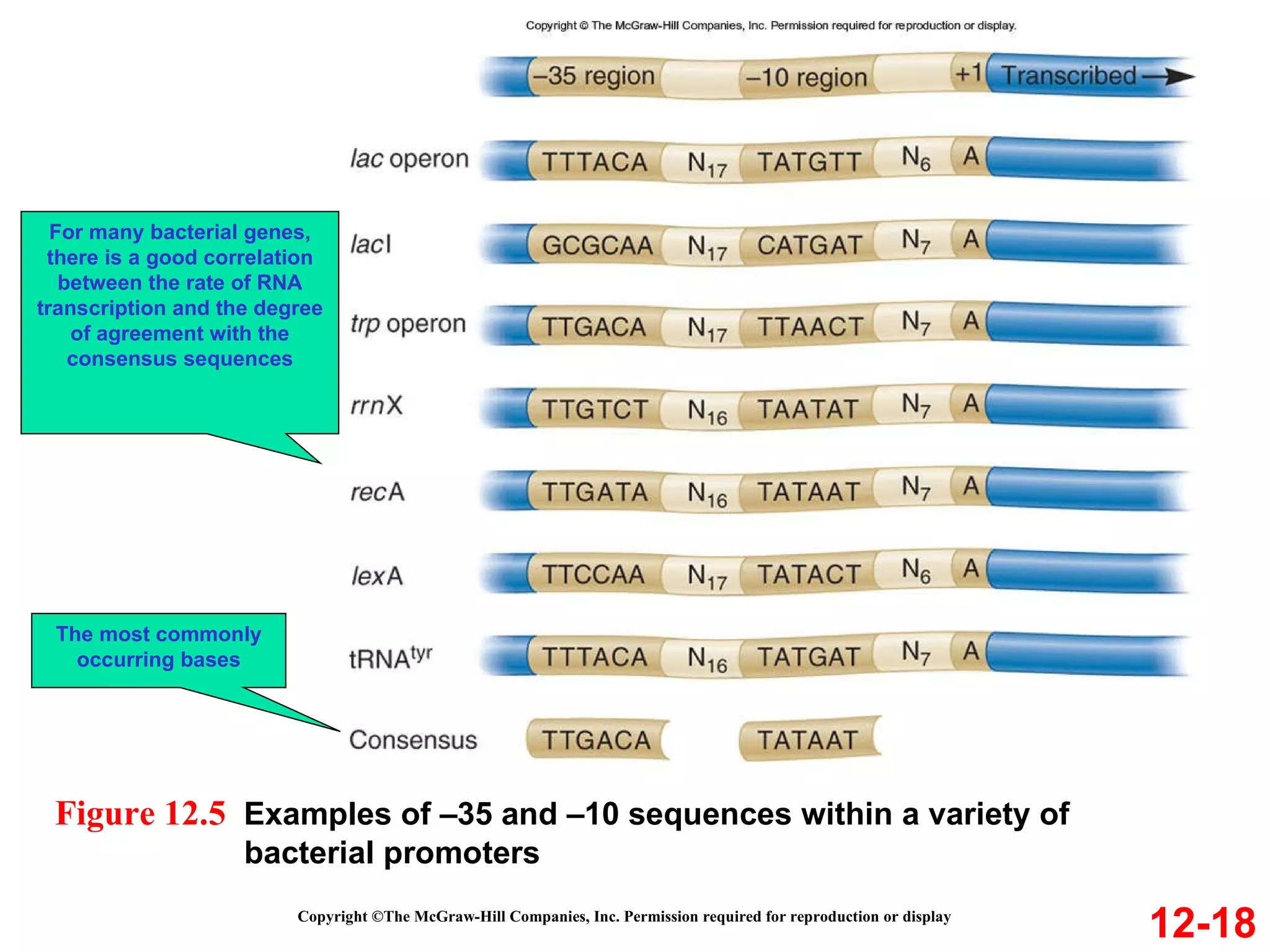
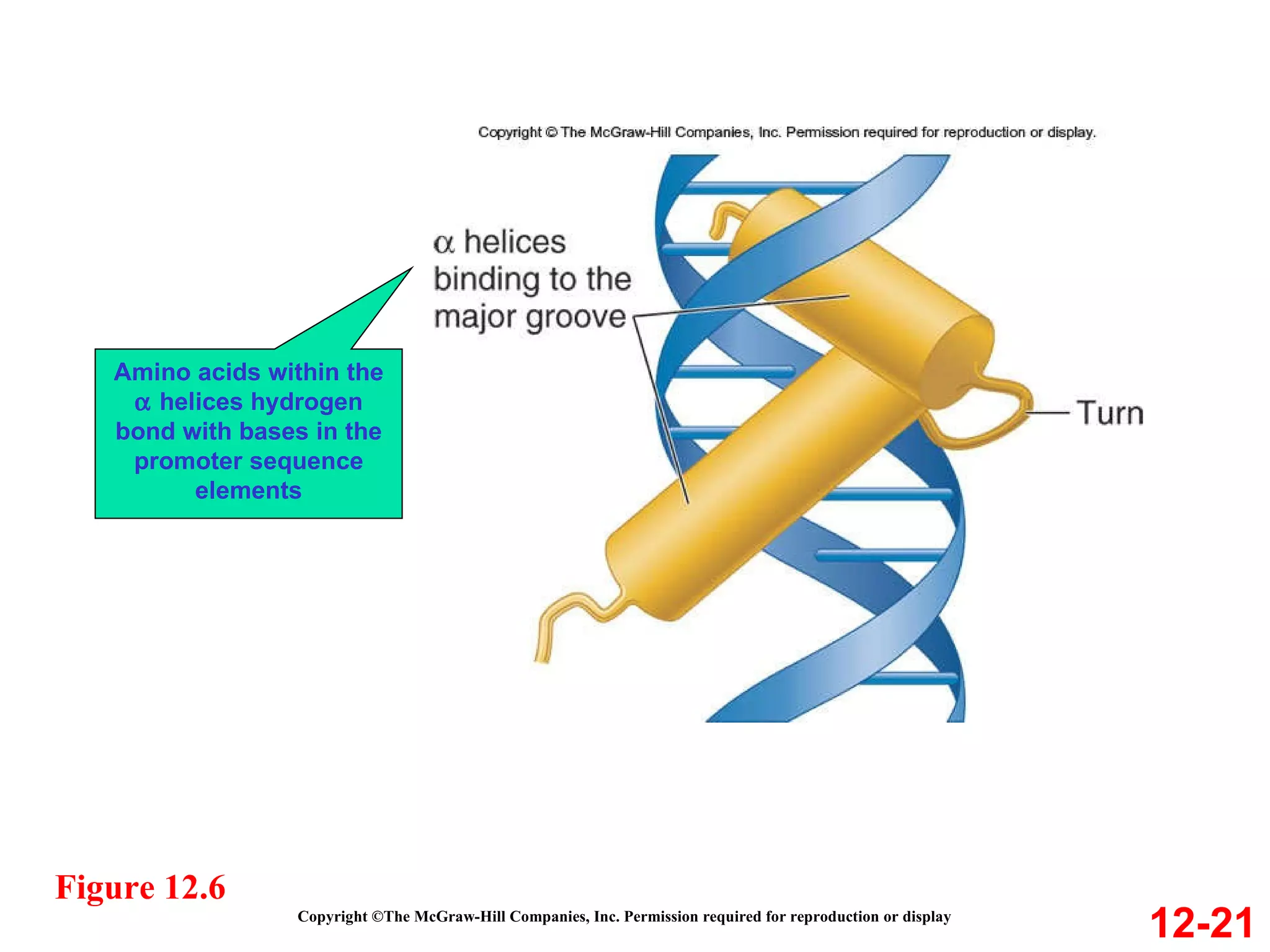
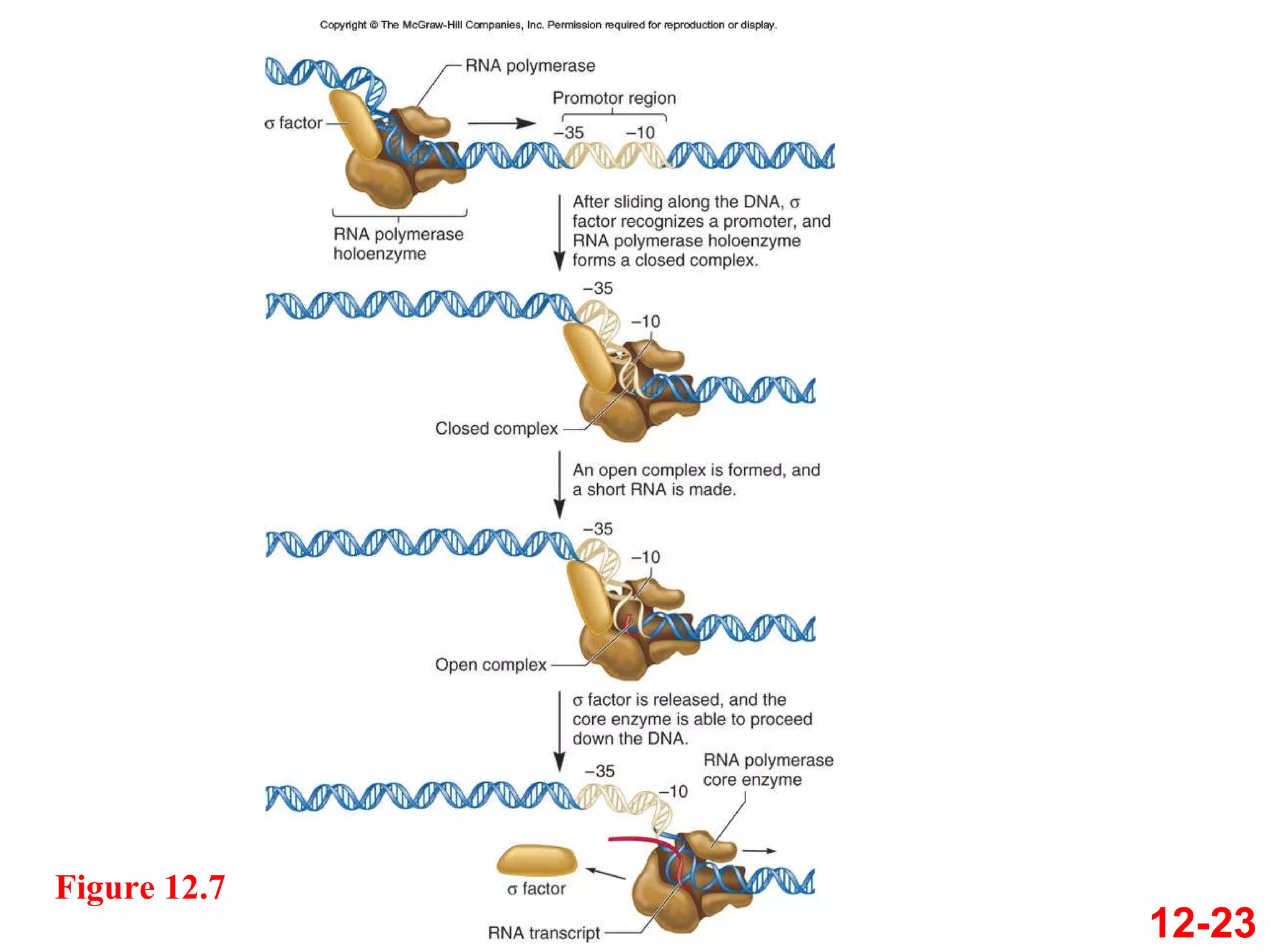
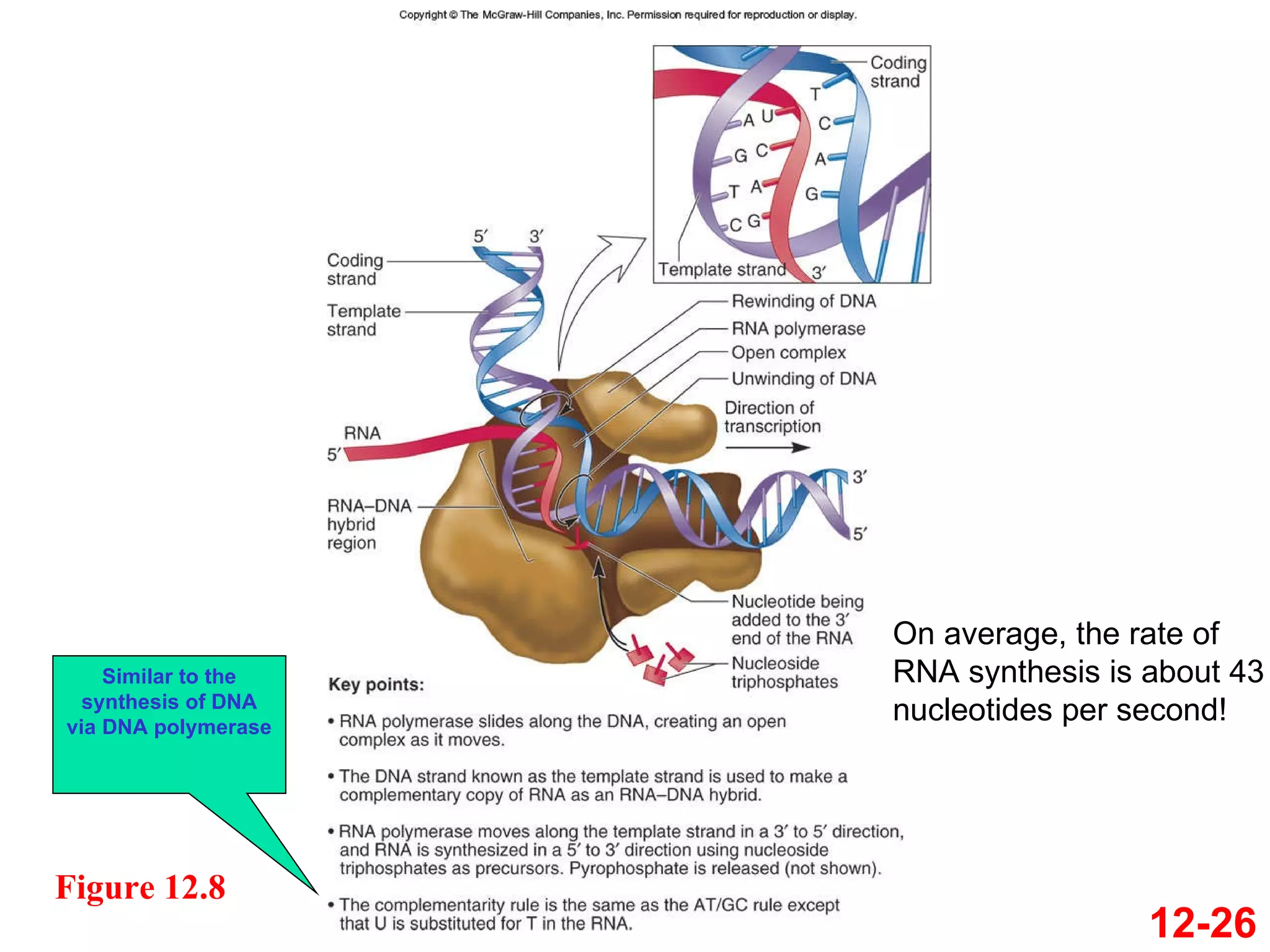
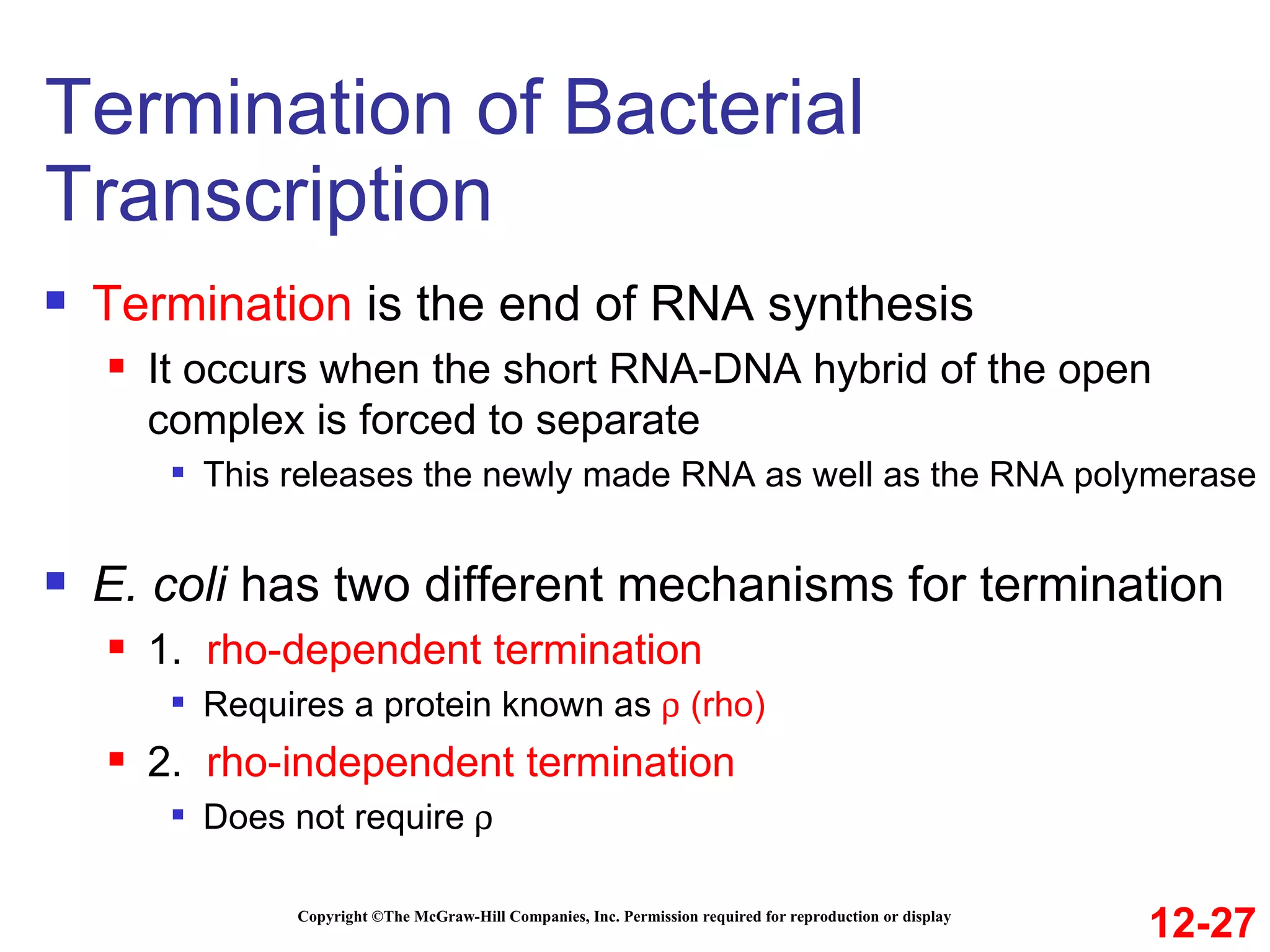
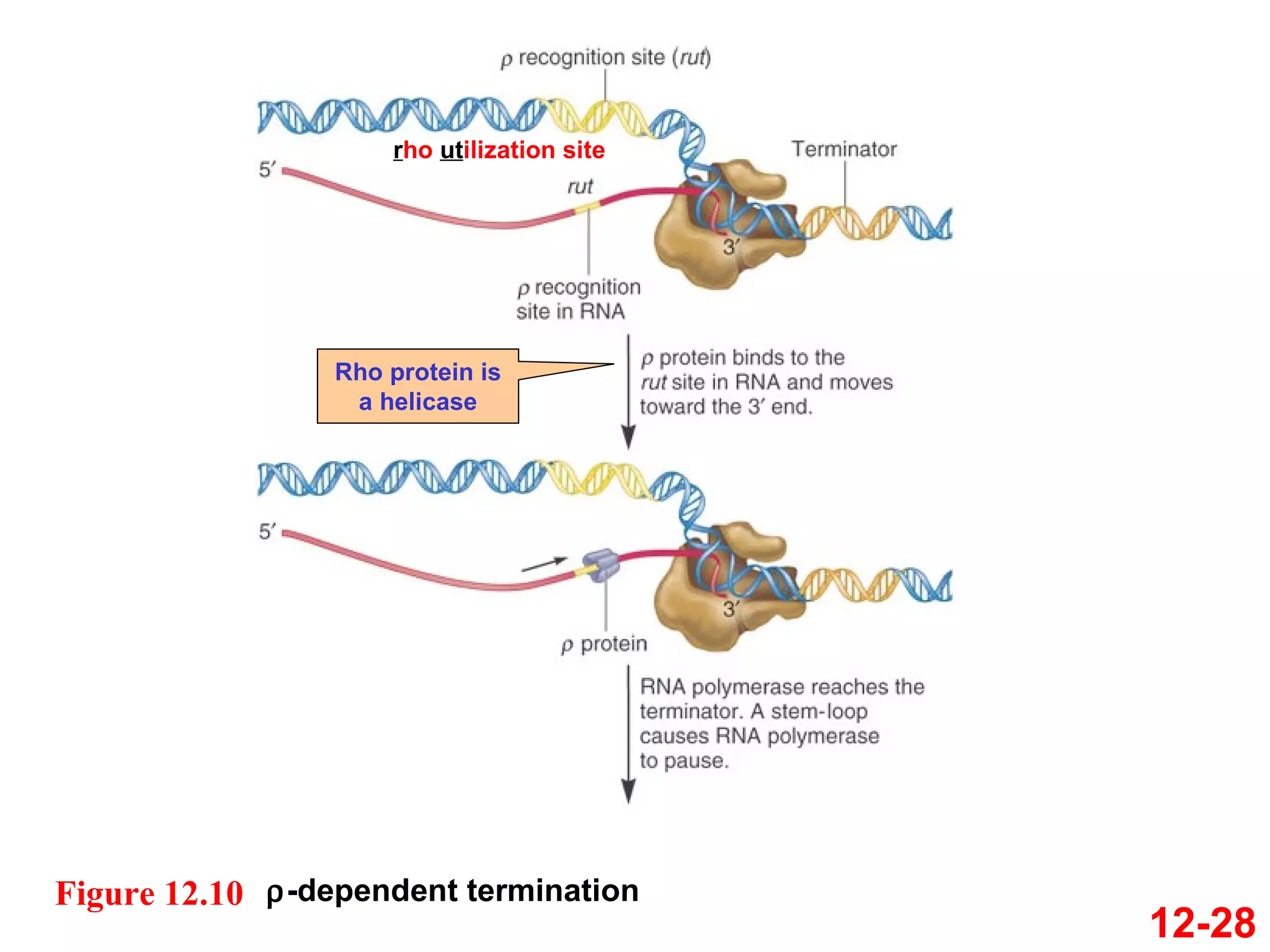
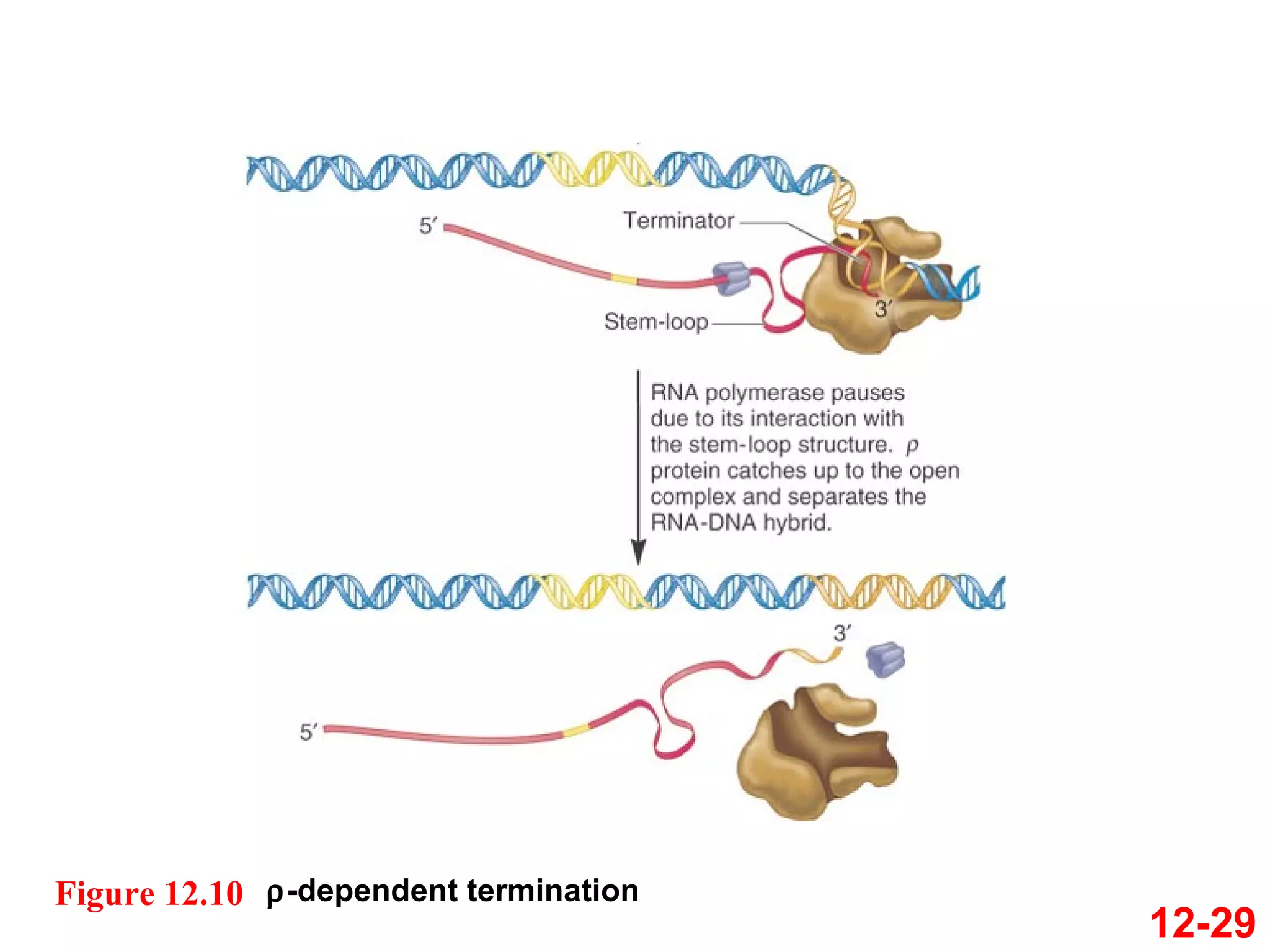
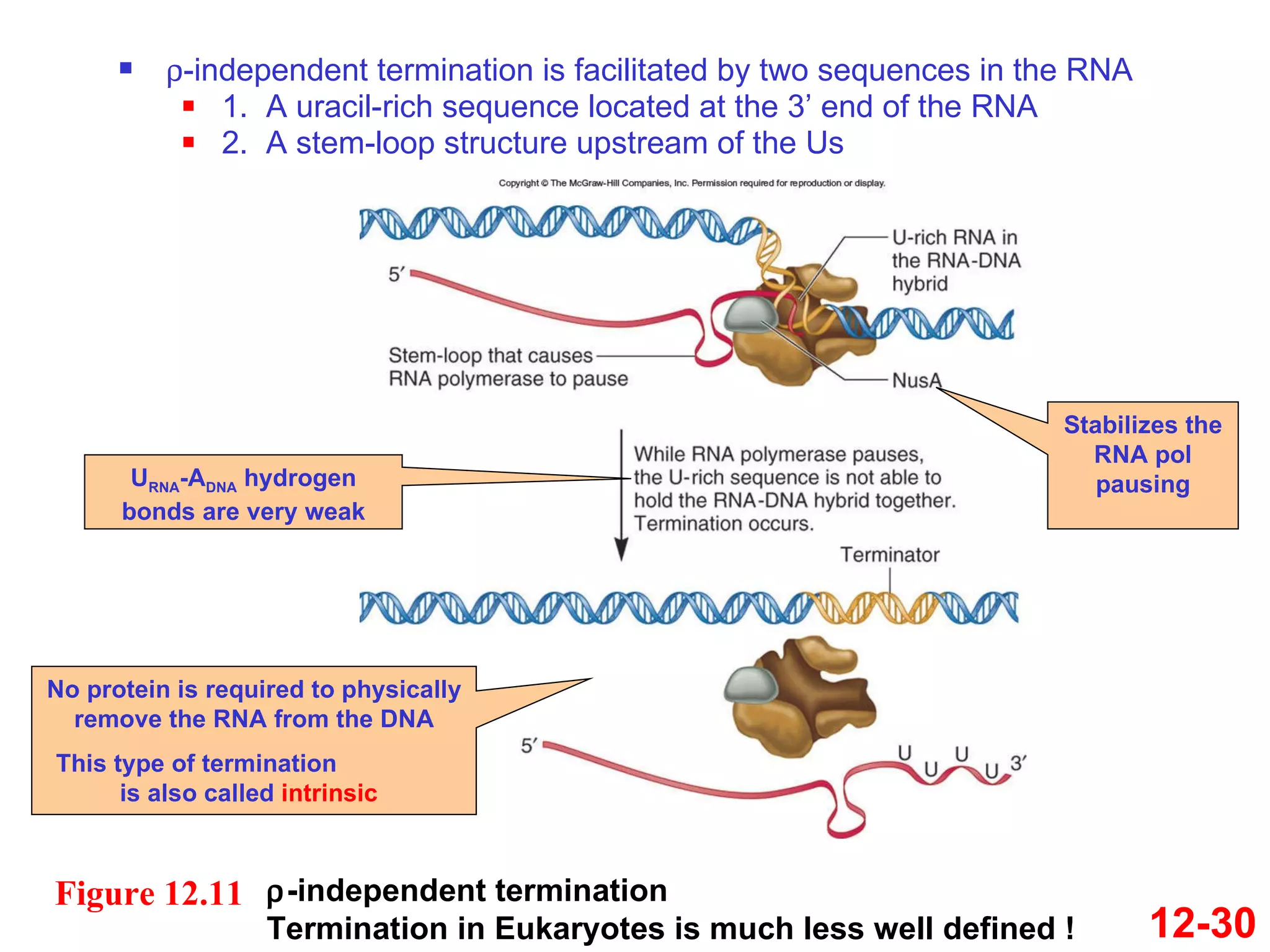
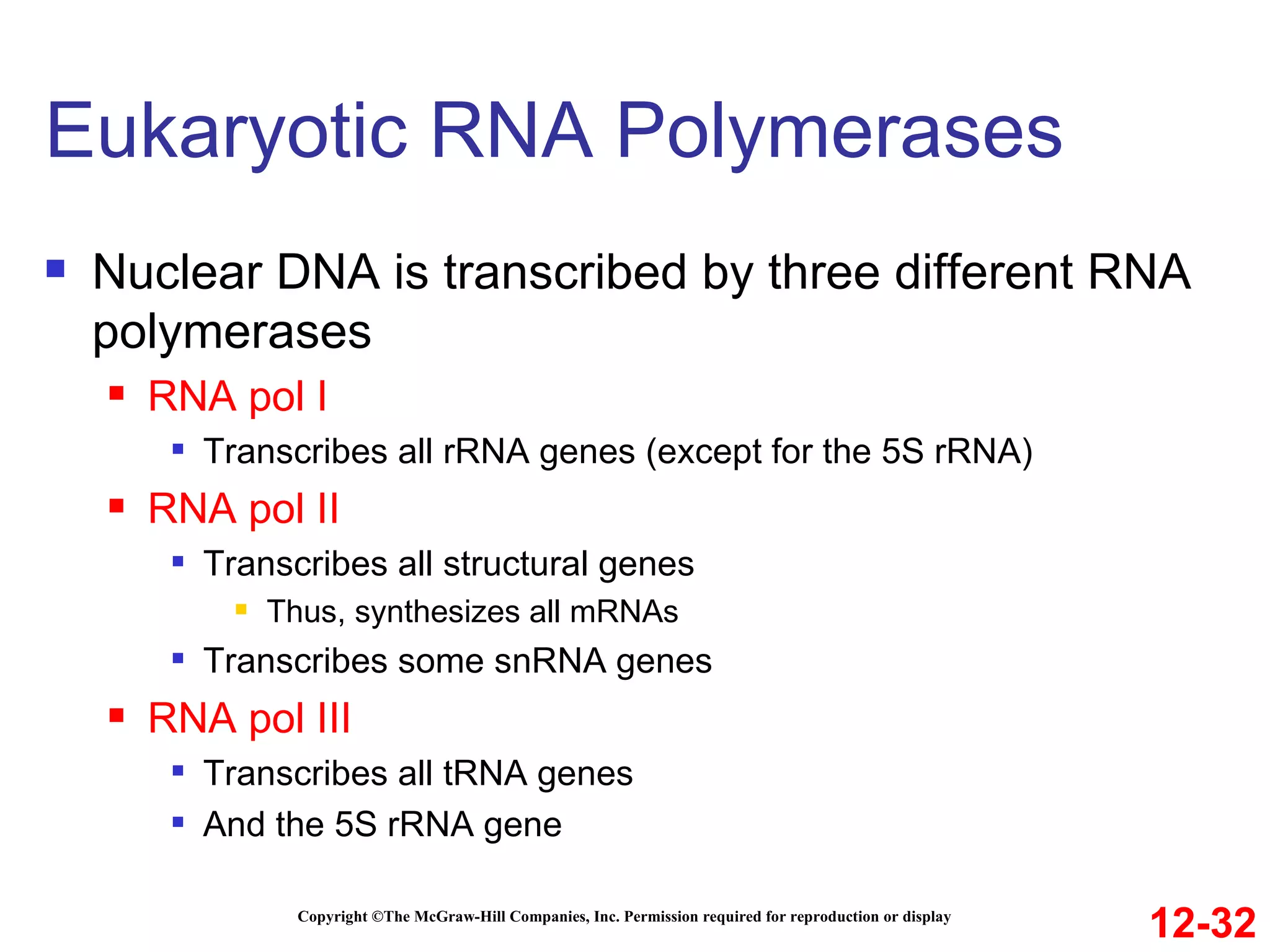
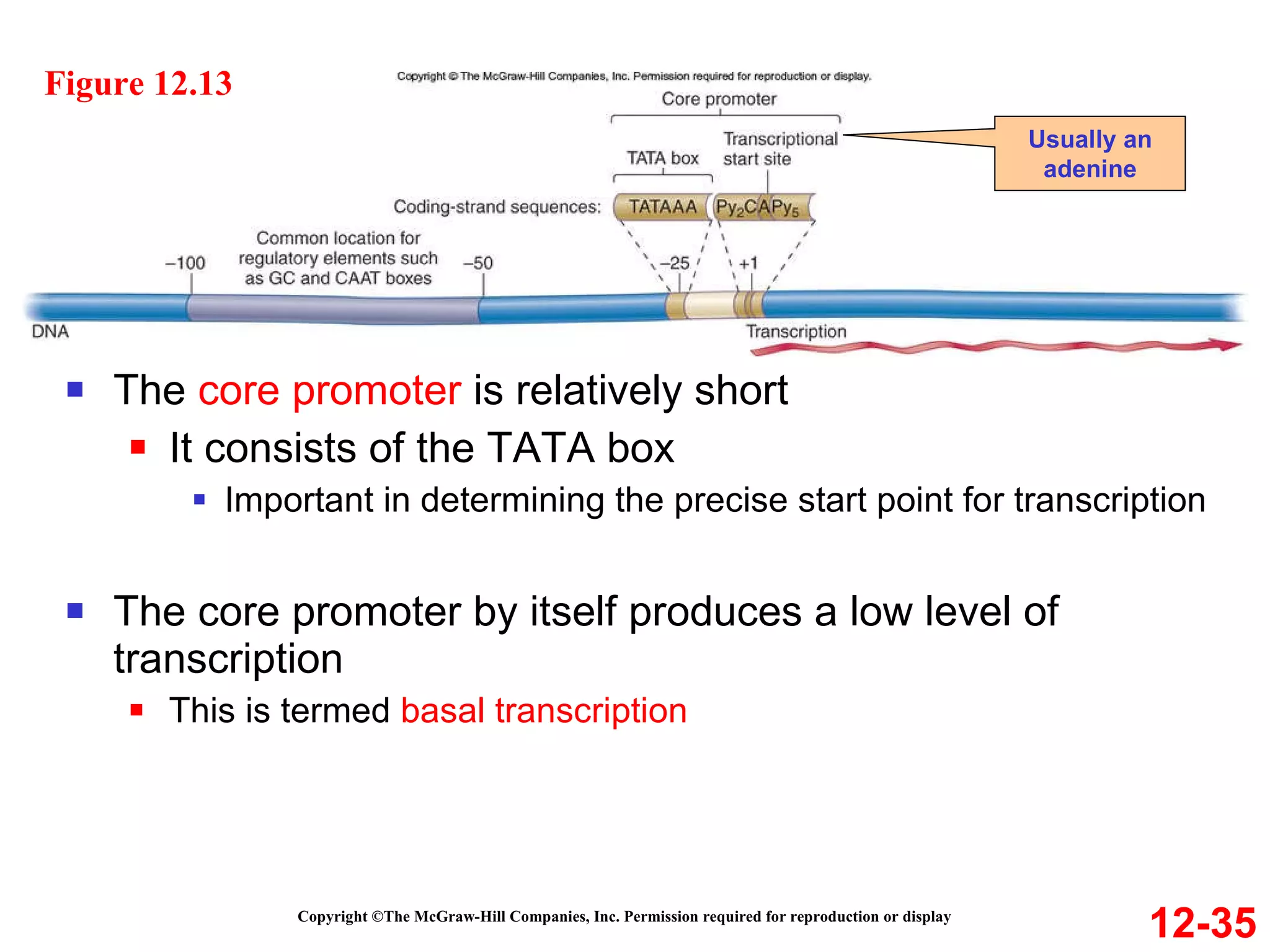
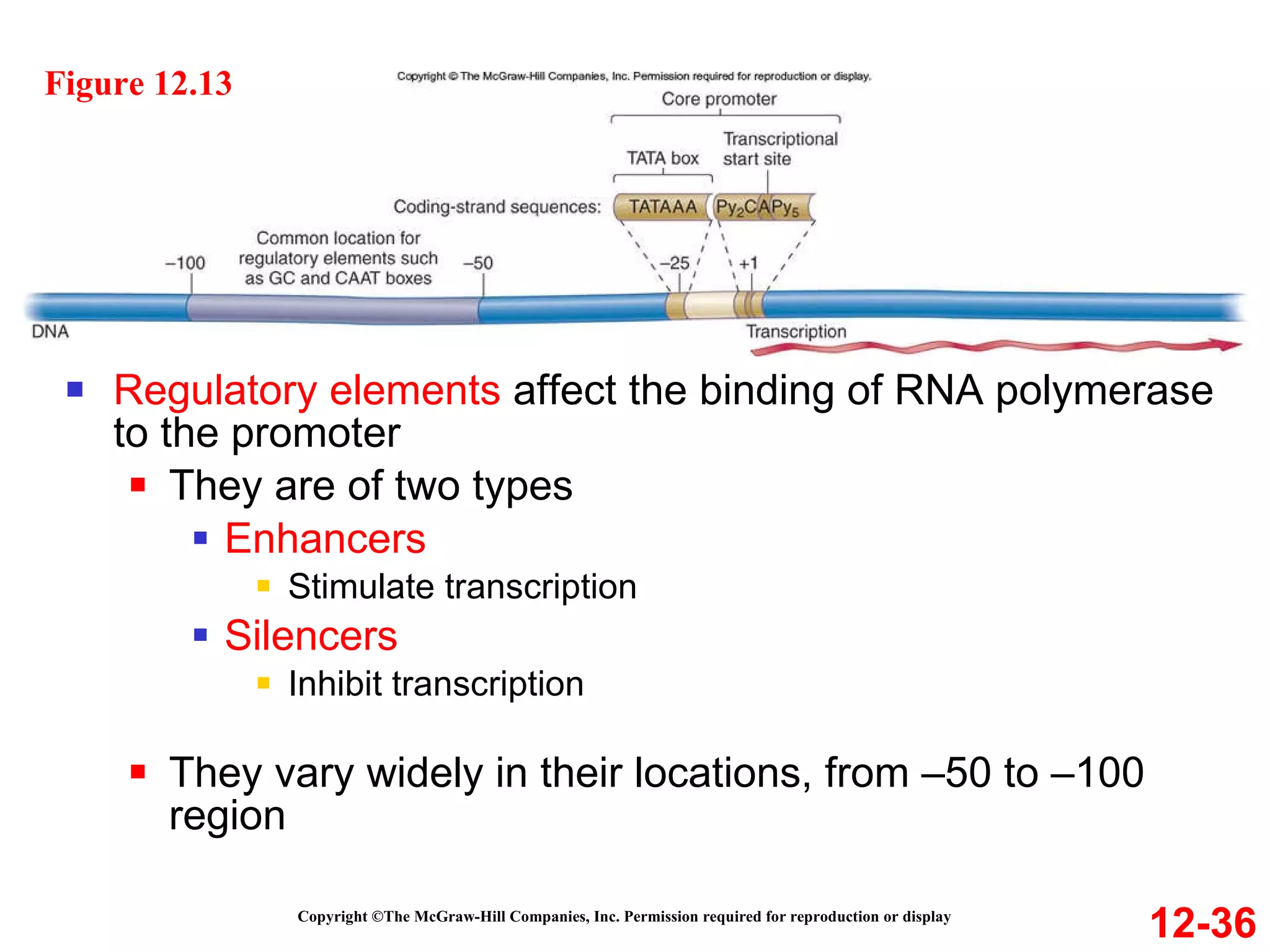
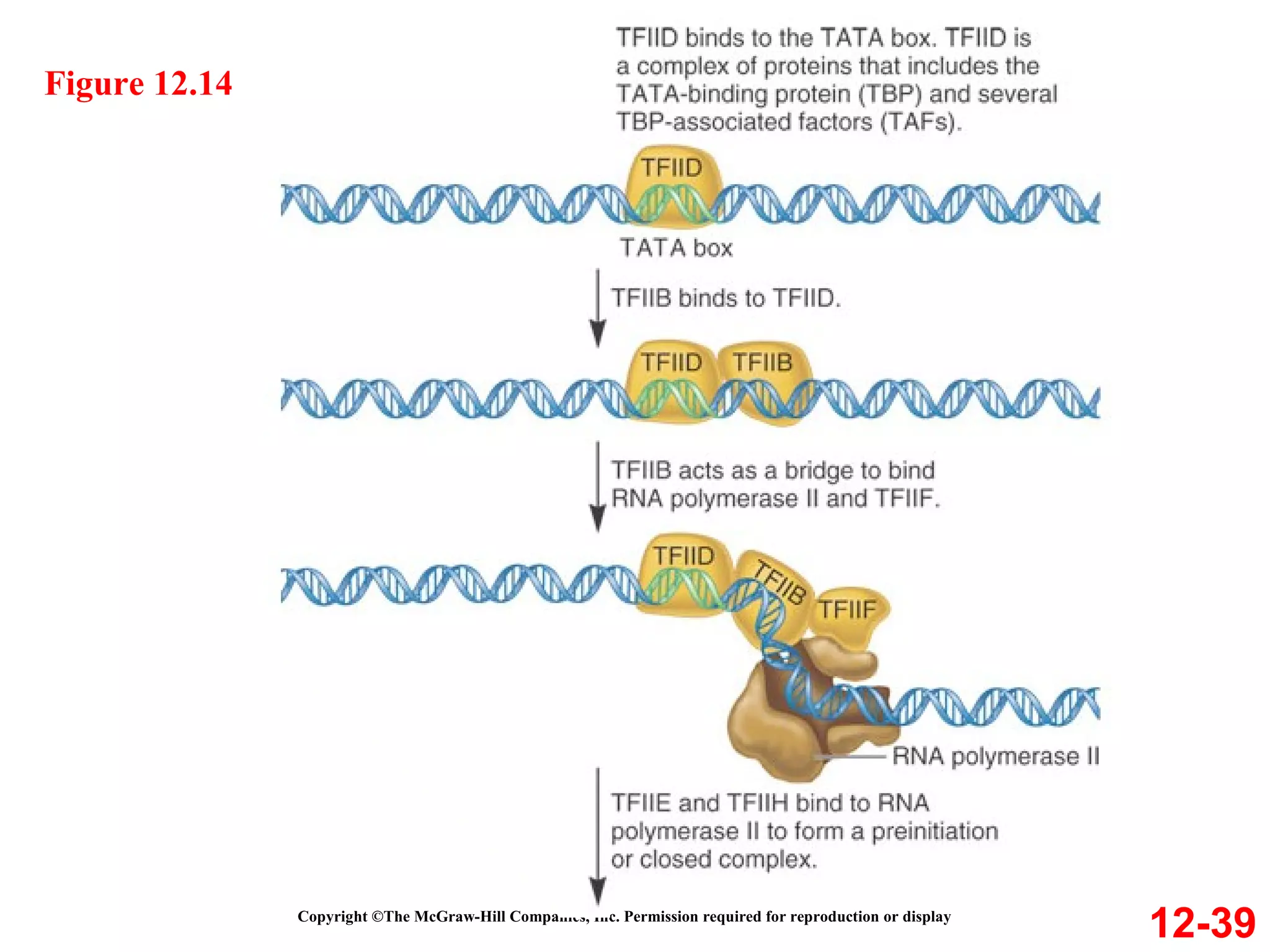
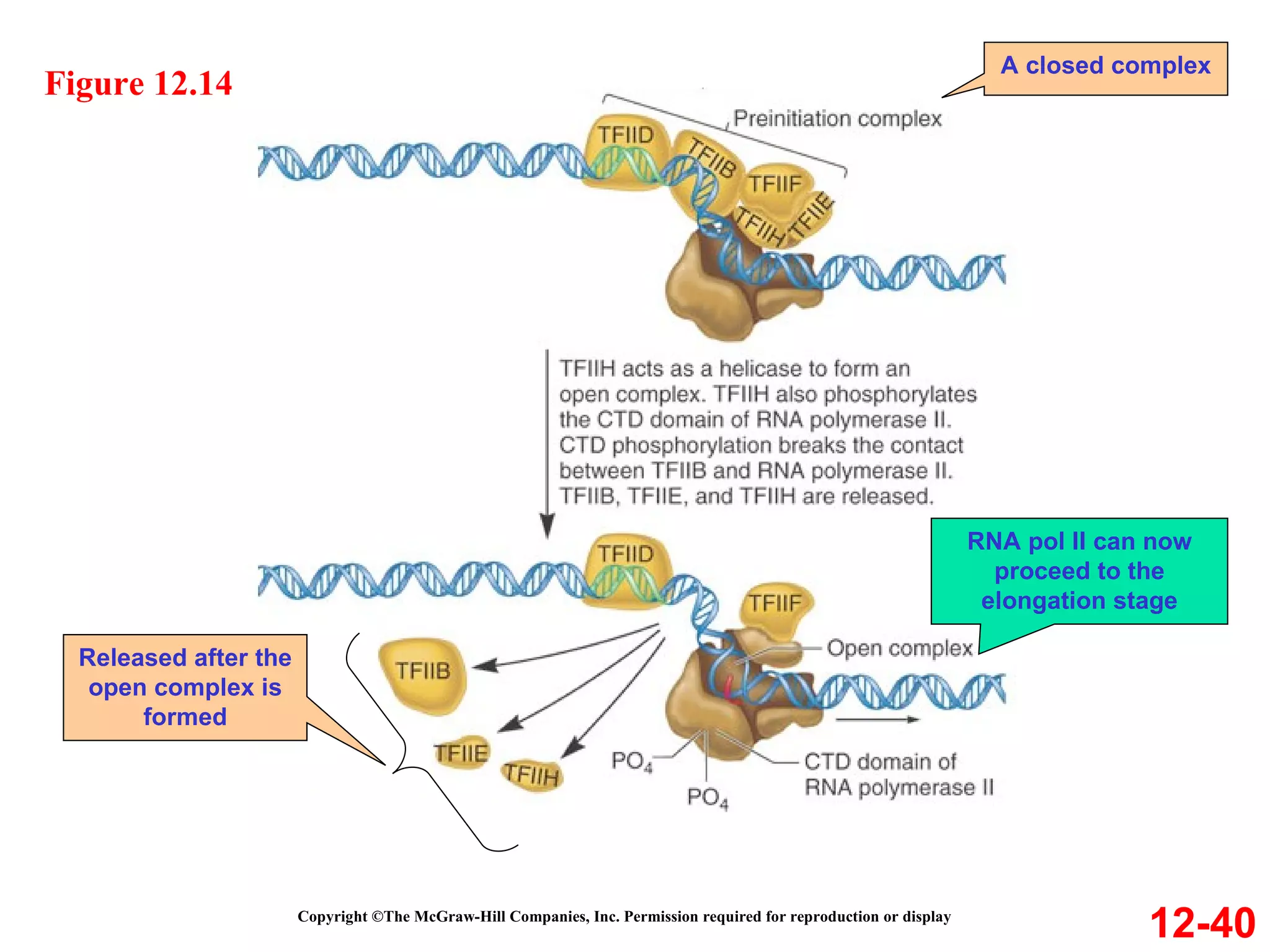
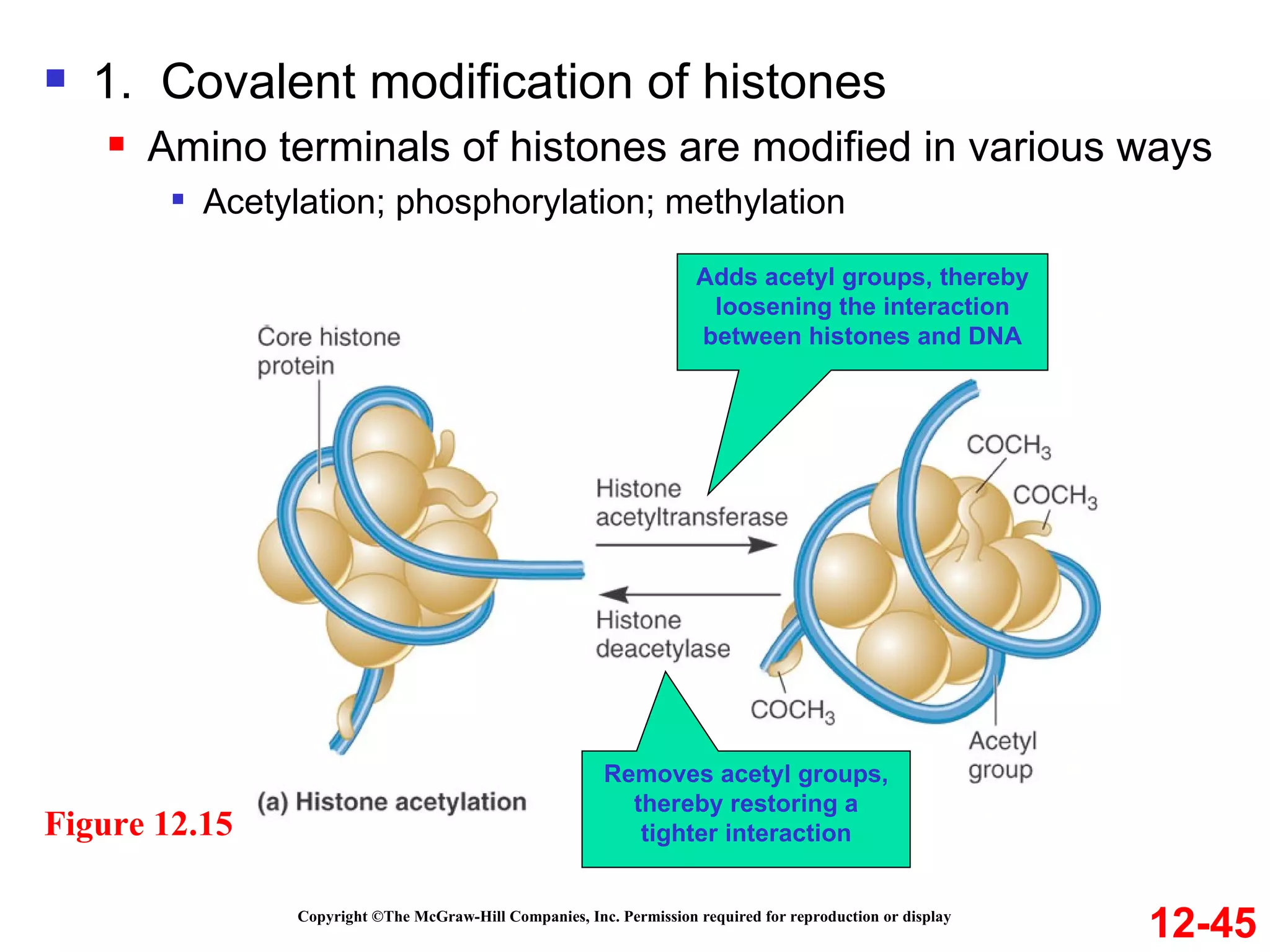
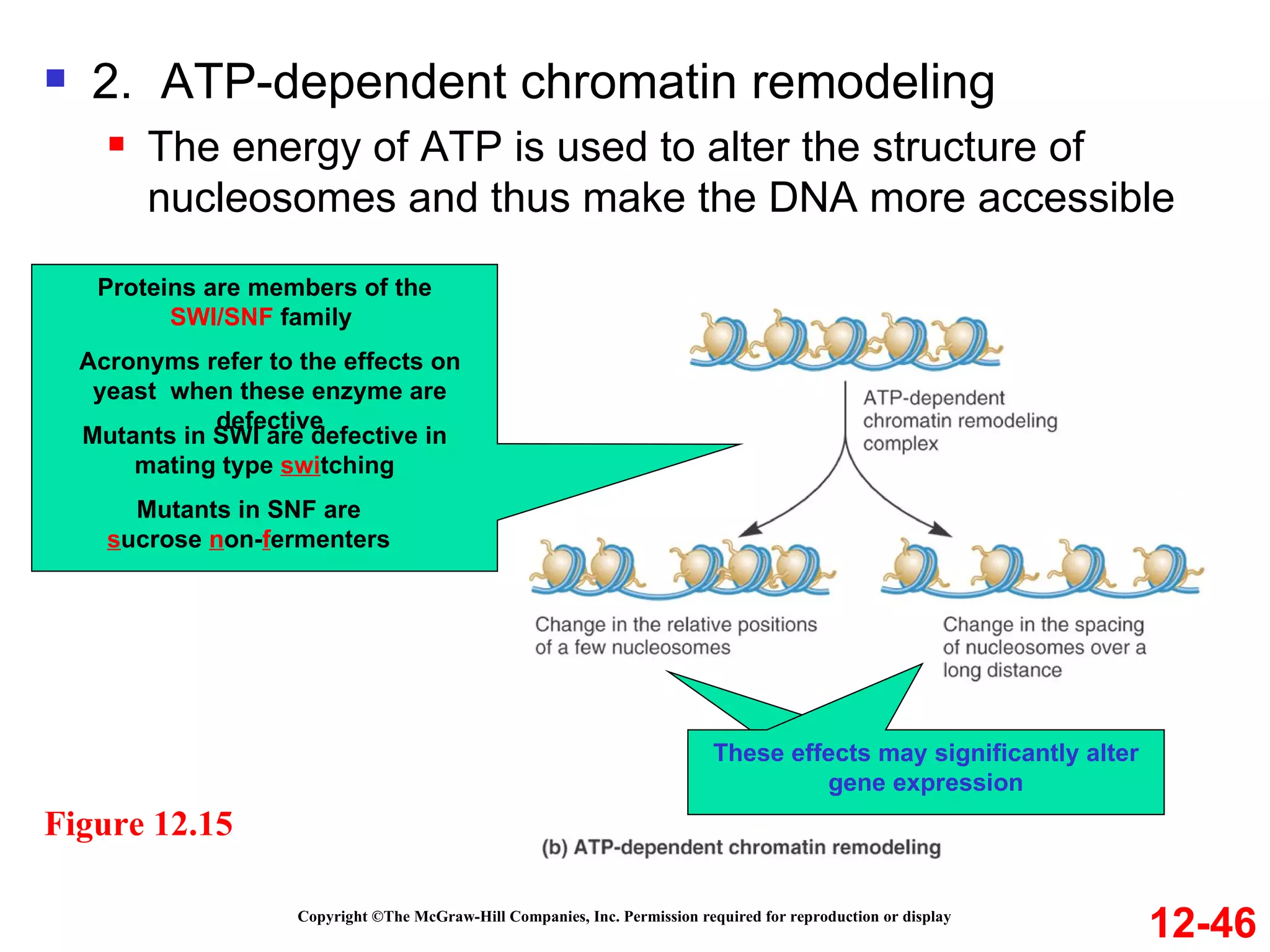
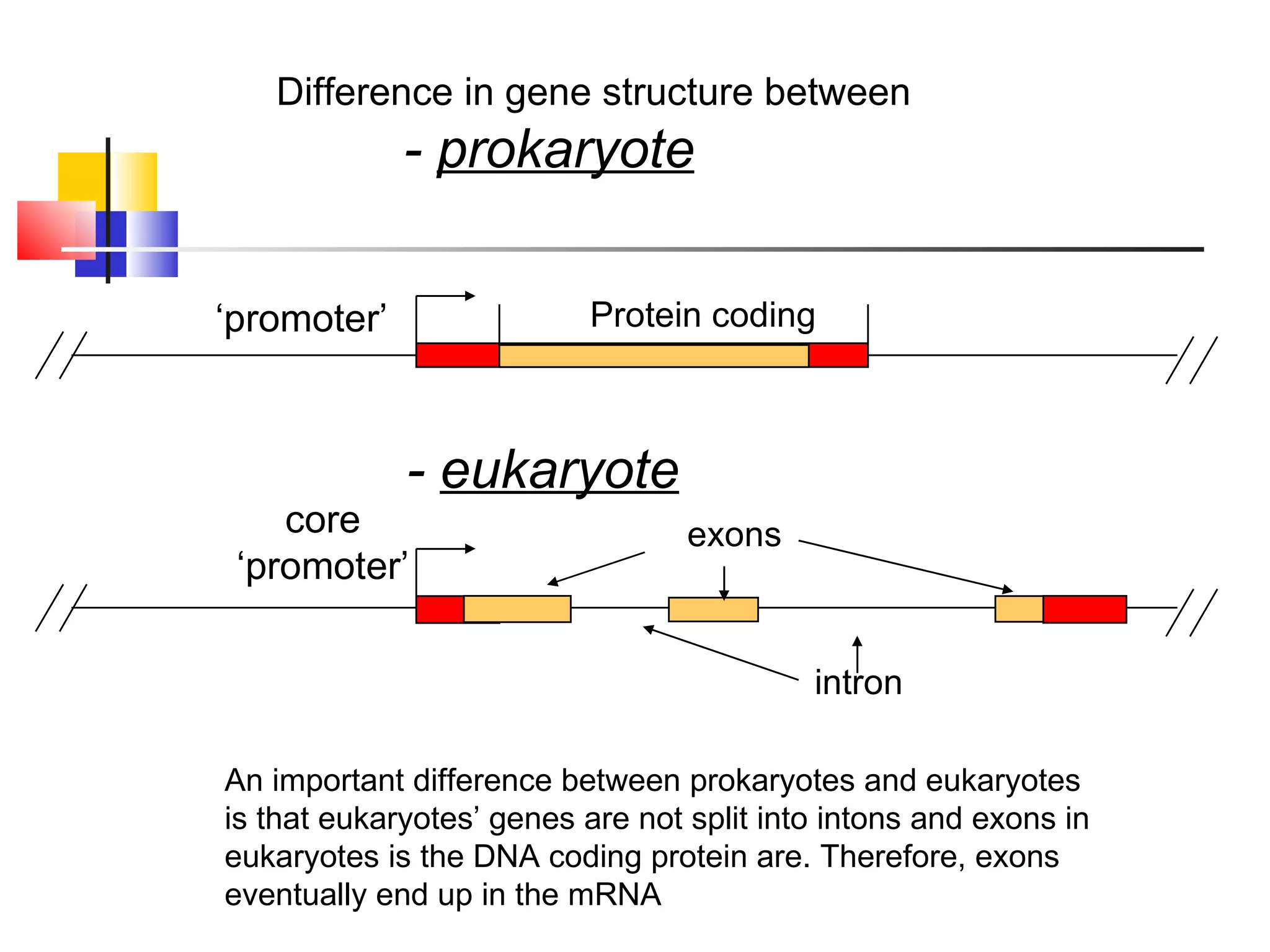
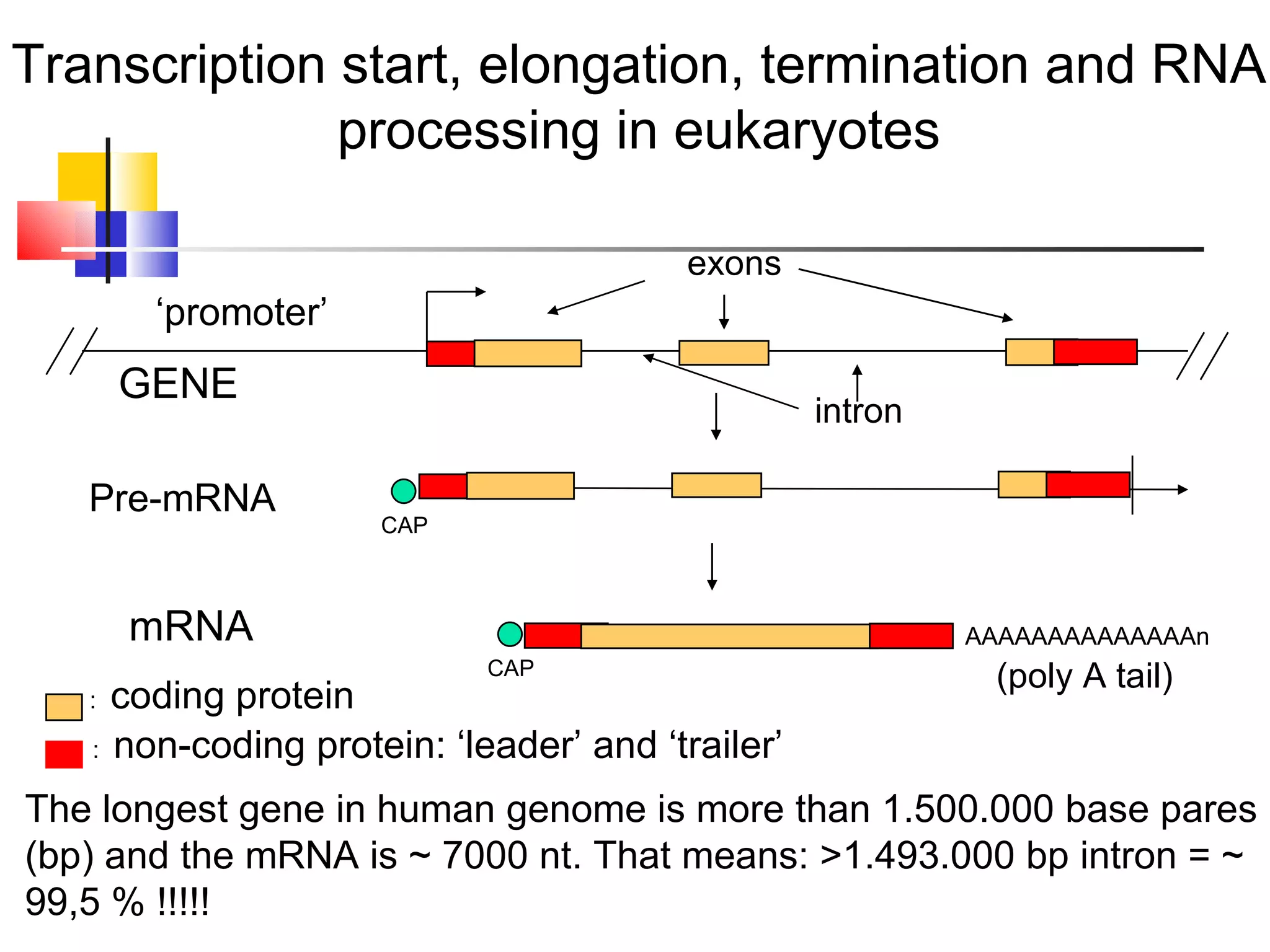
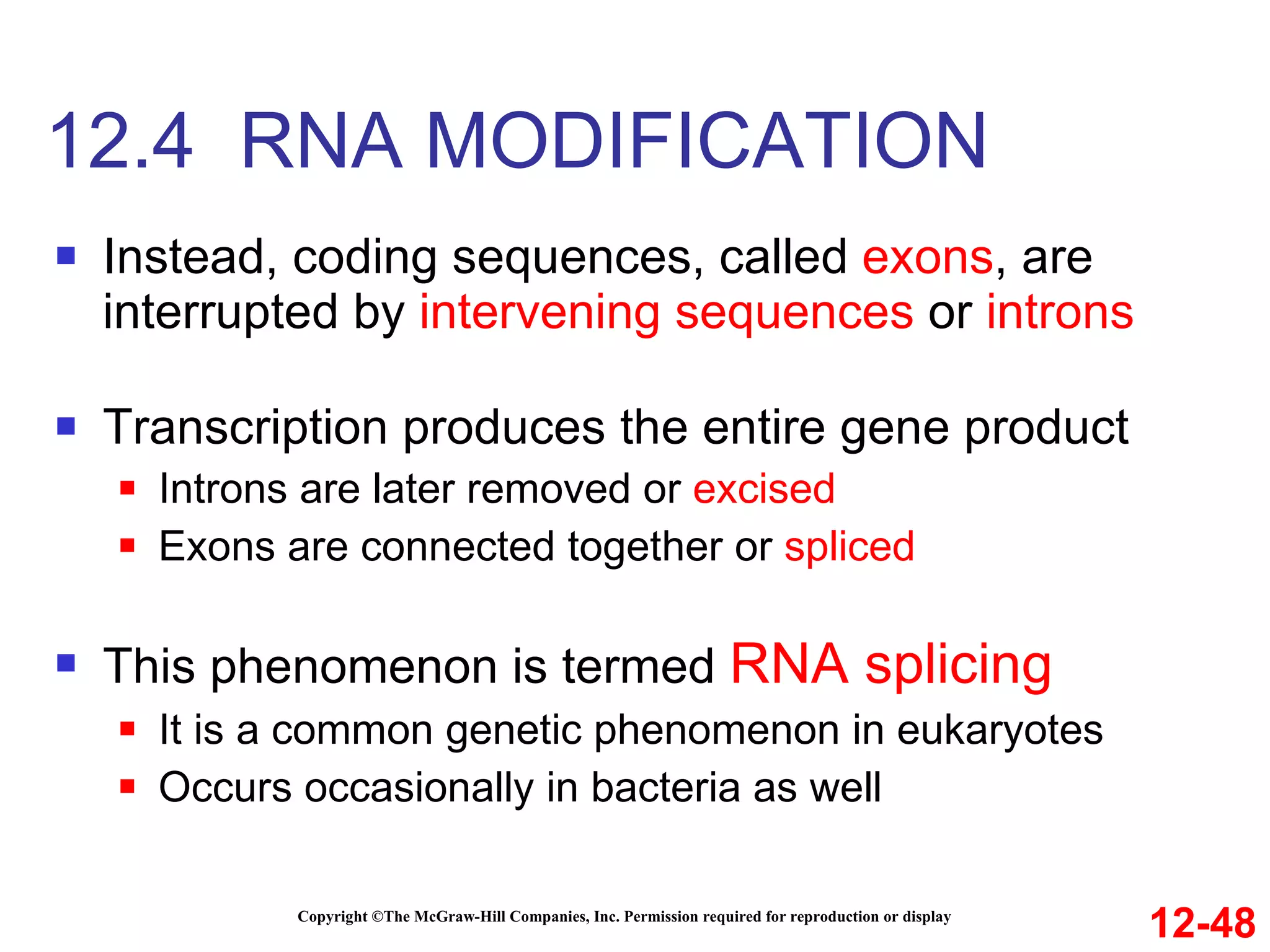
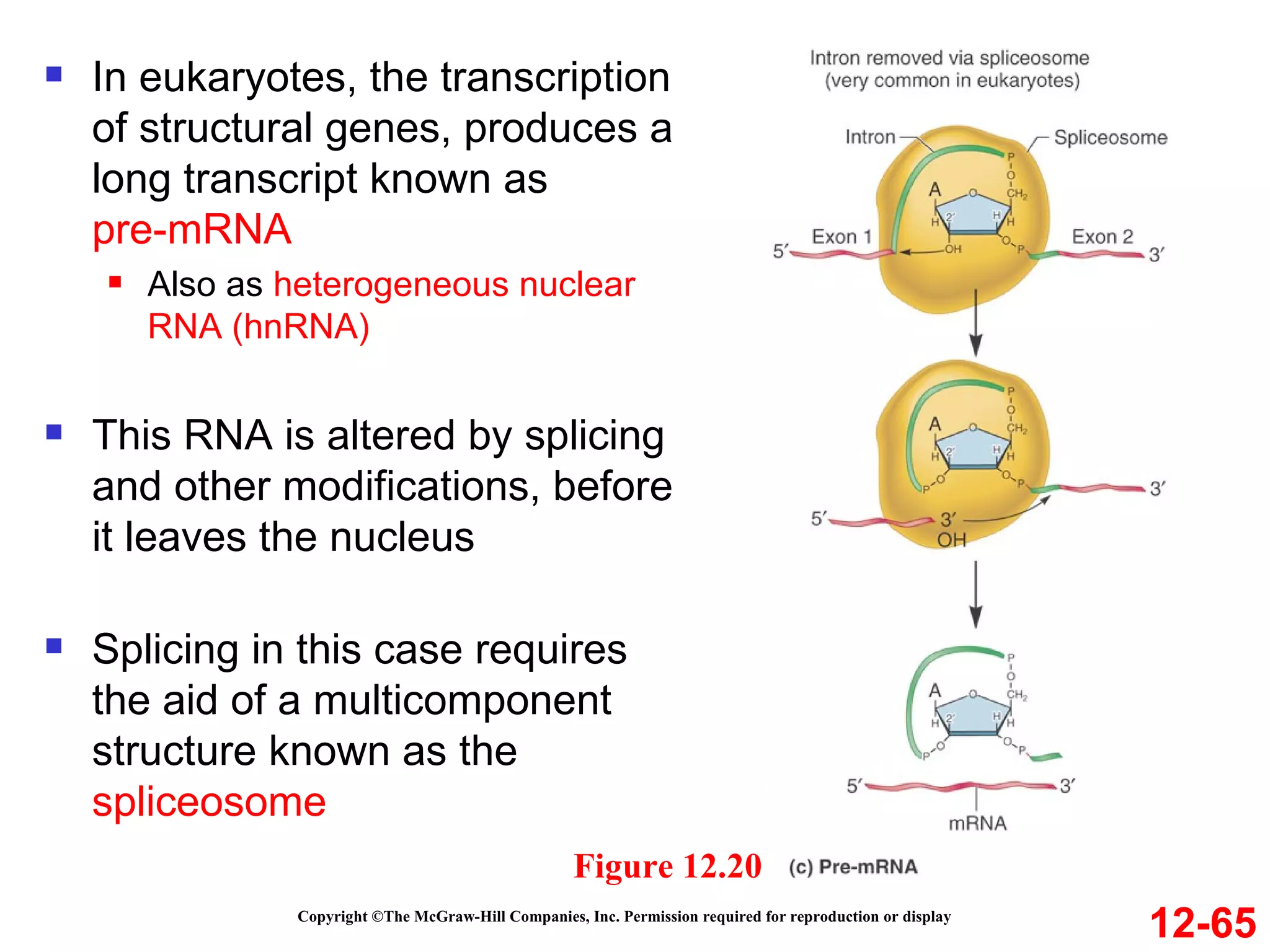
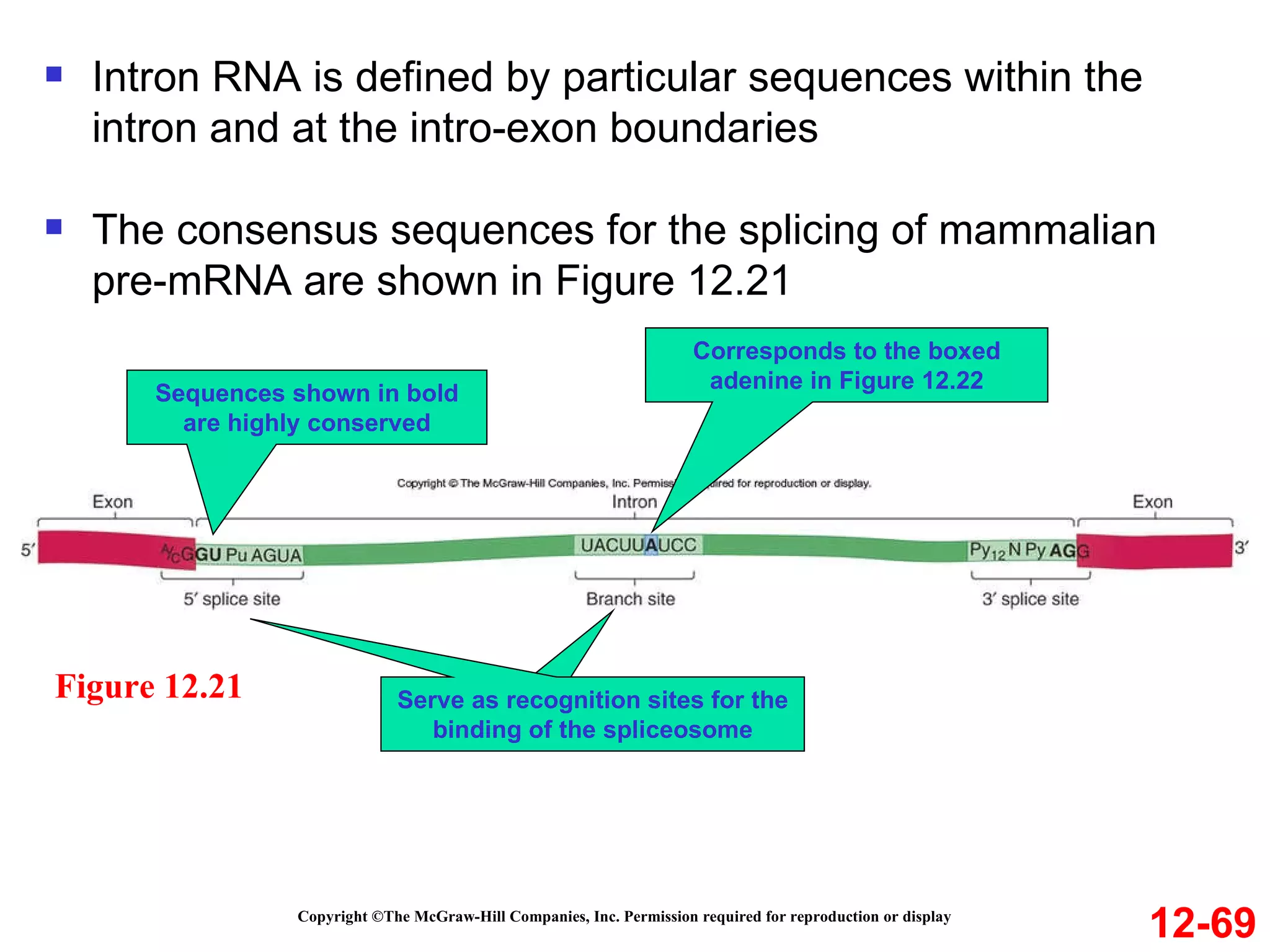
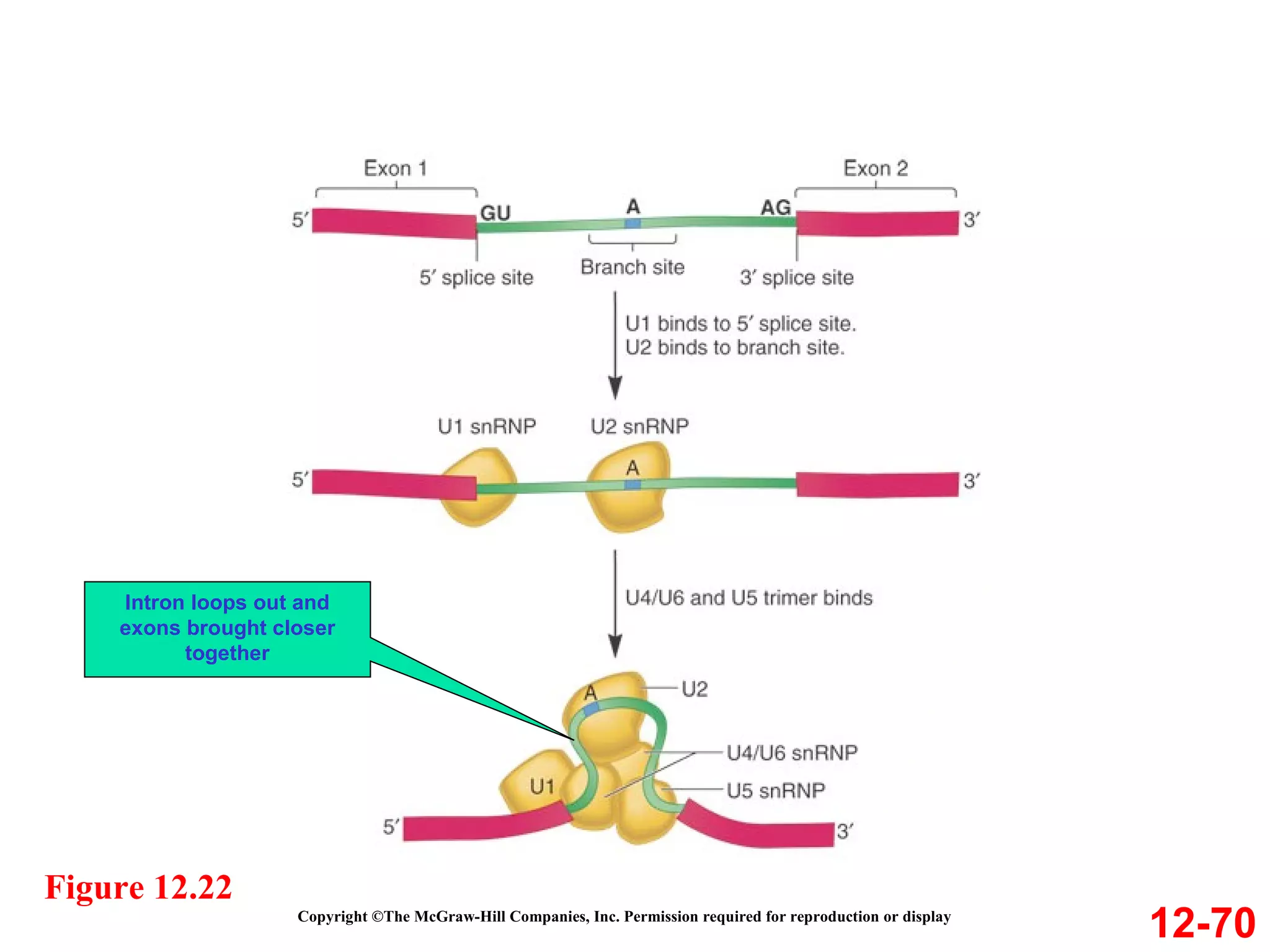
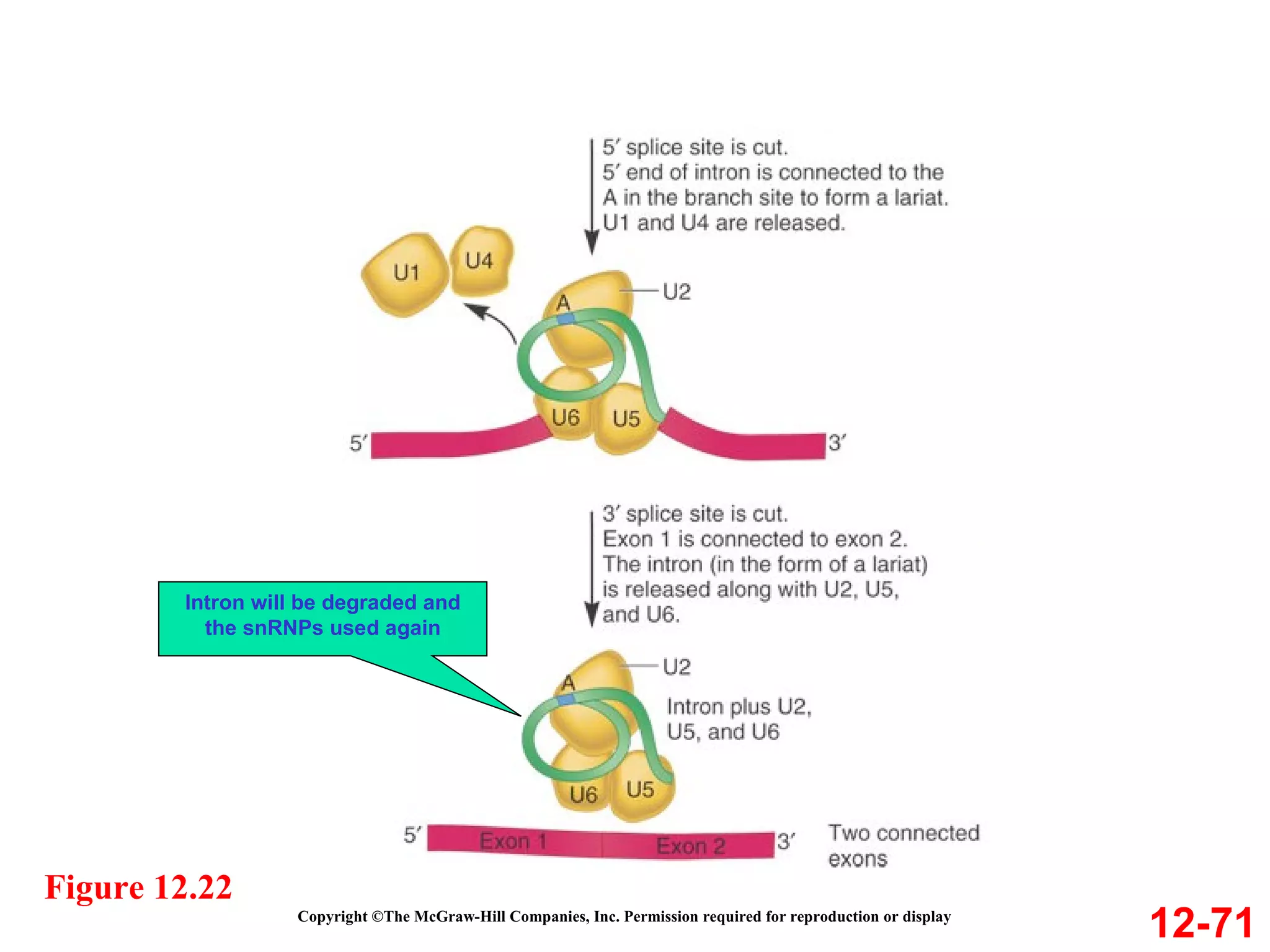
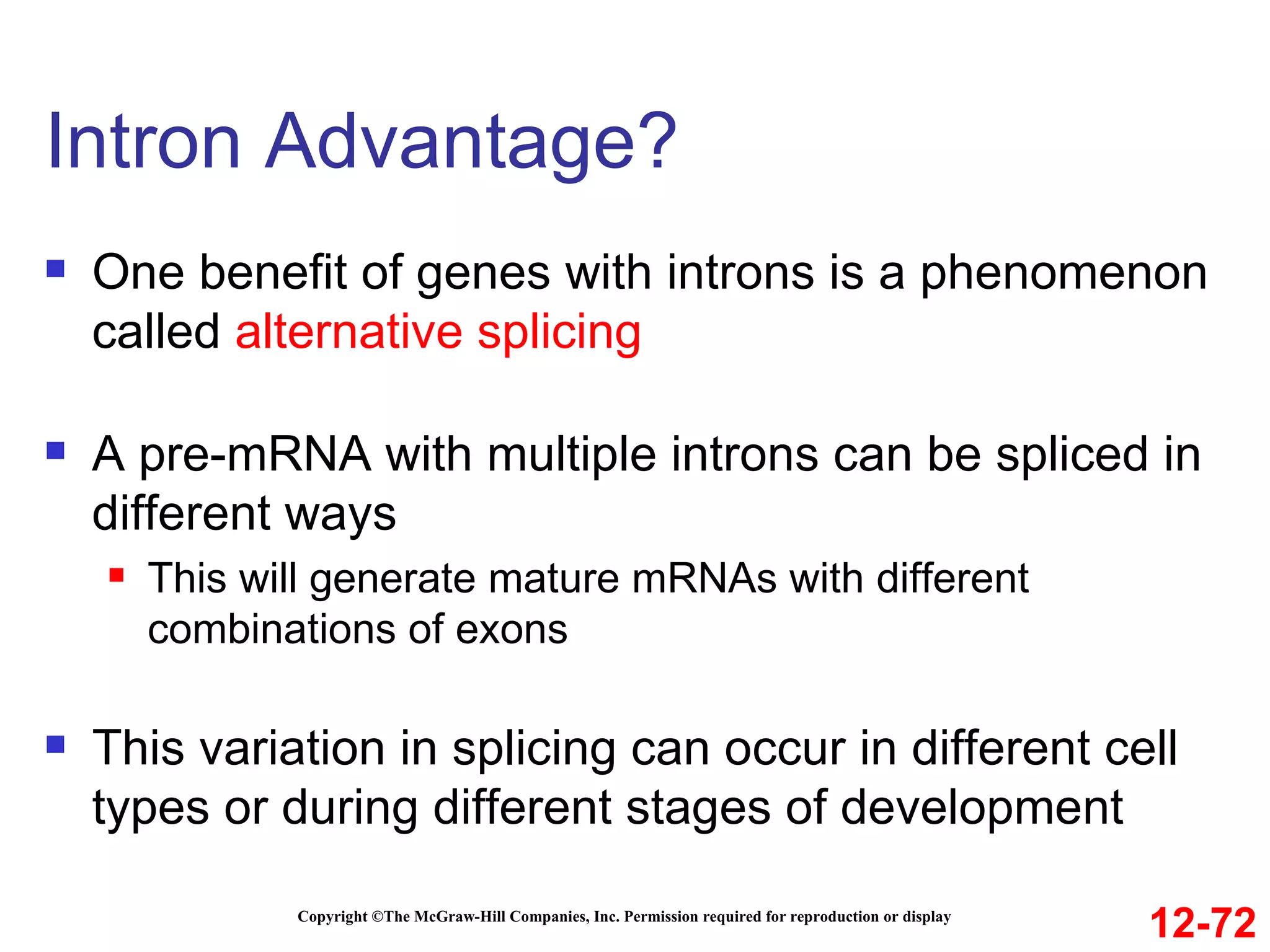
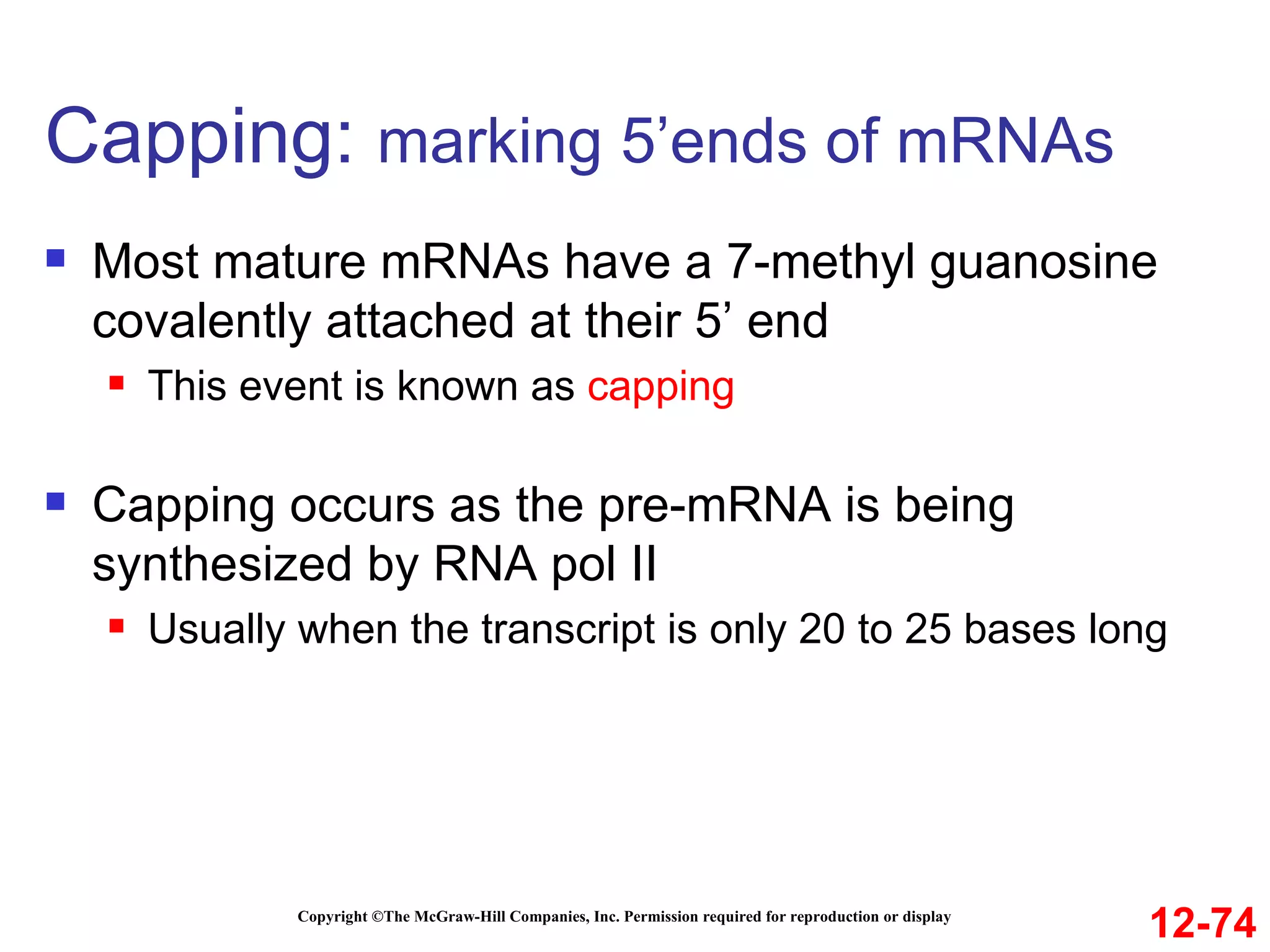
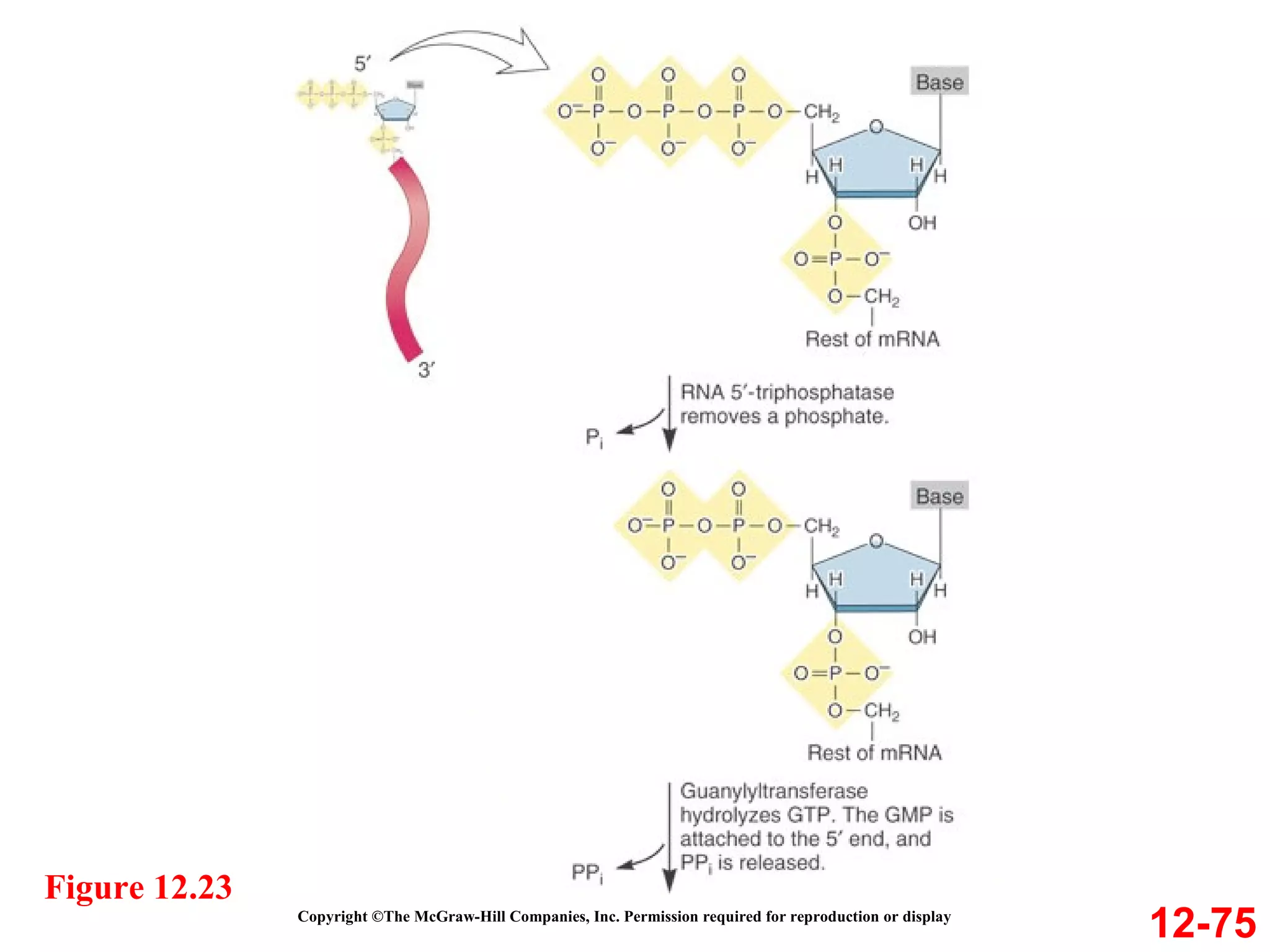
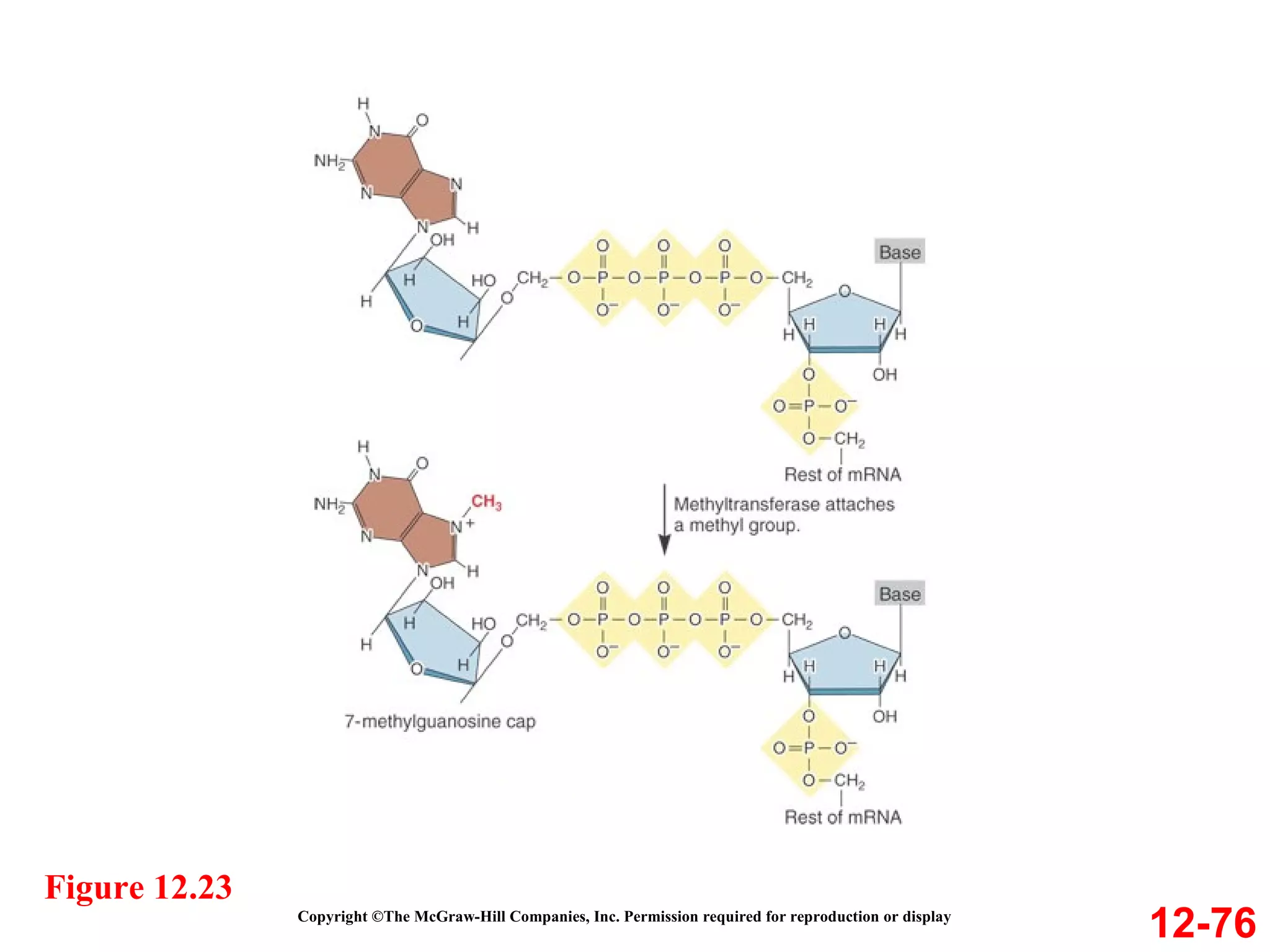
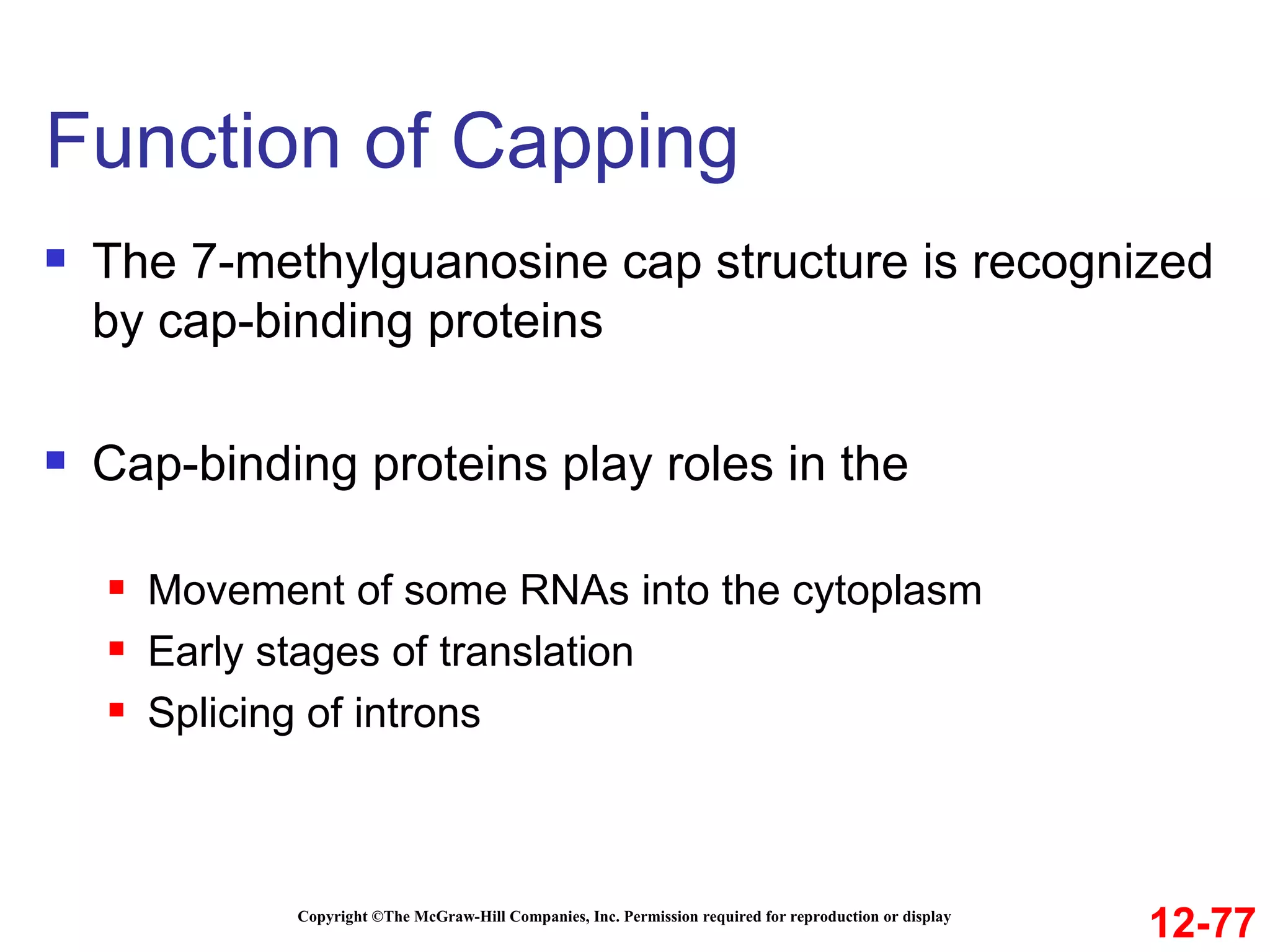
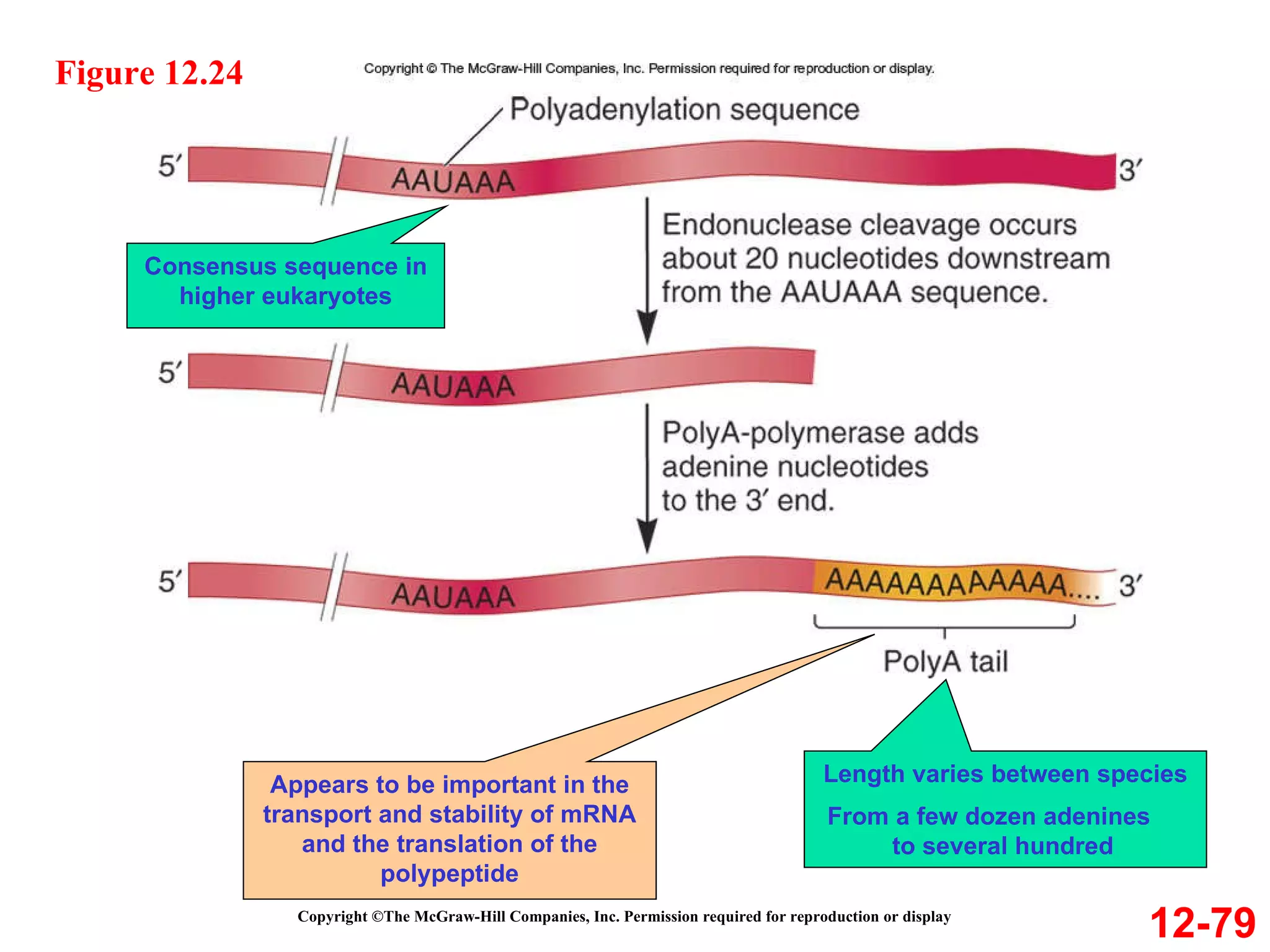
The document discusses gene transcription and RNA processing in bacteria and eukaryotes. It covers the key steps and mechanisms of transcription, including promoters, initiation, elongation, and termination. In eukaryotes, transcription is more complex due to larger genomes, cellular complexity, and need for precise gene regulation in different cell types. The three classes of RNA polymerases and their roles are described, as well as sequences involved in eukaryotic gene promoters.