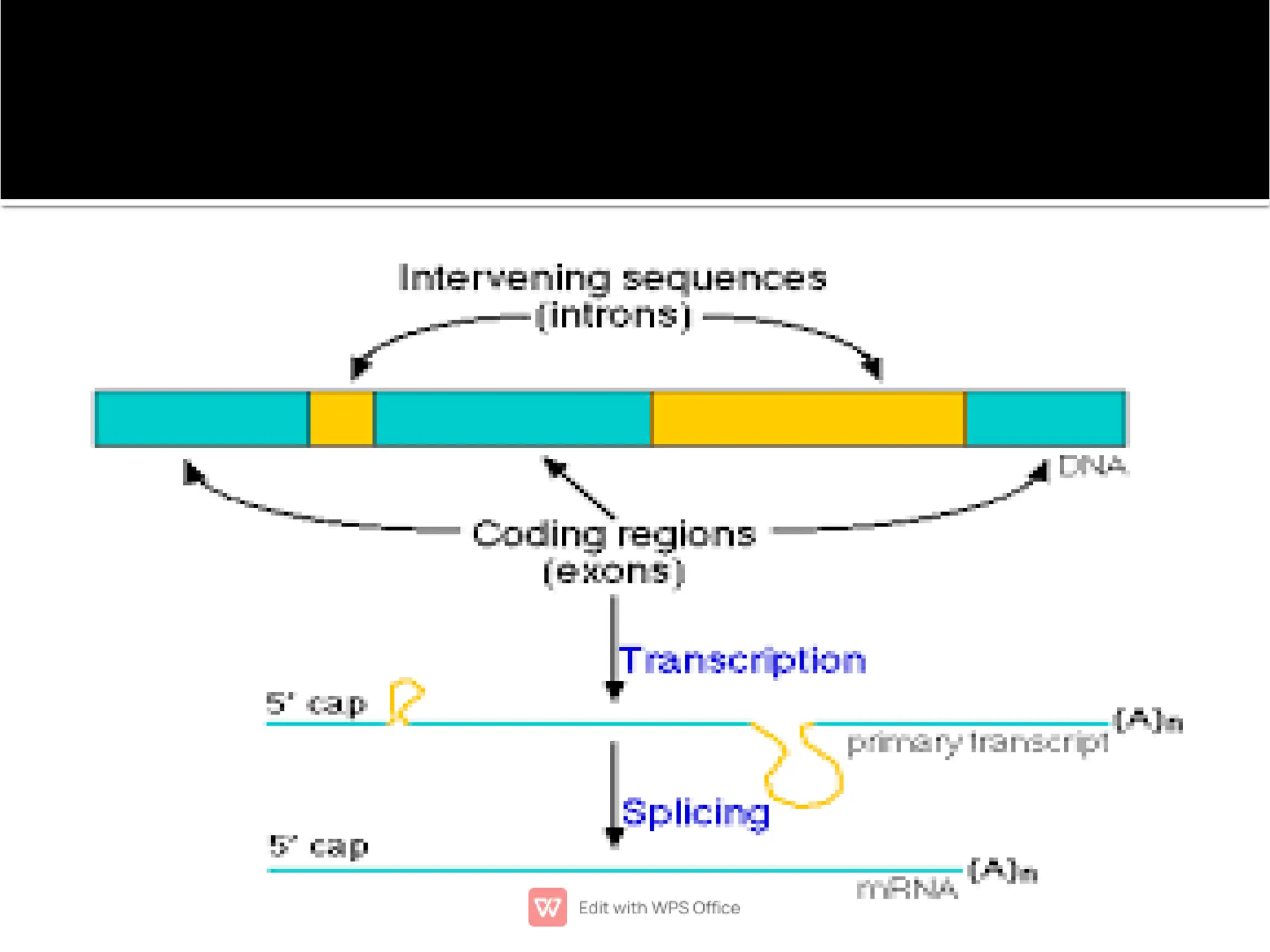
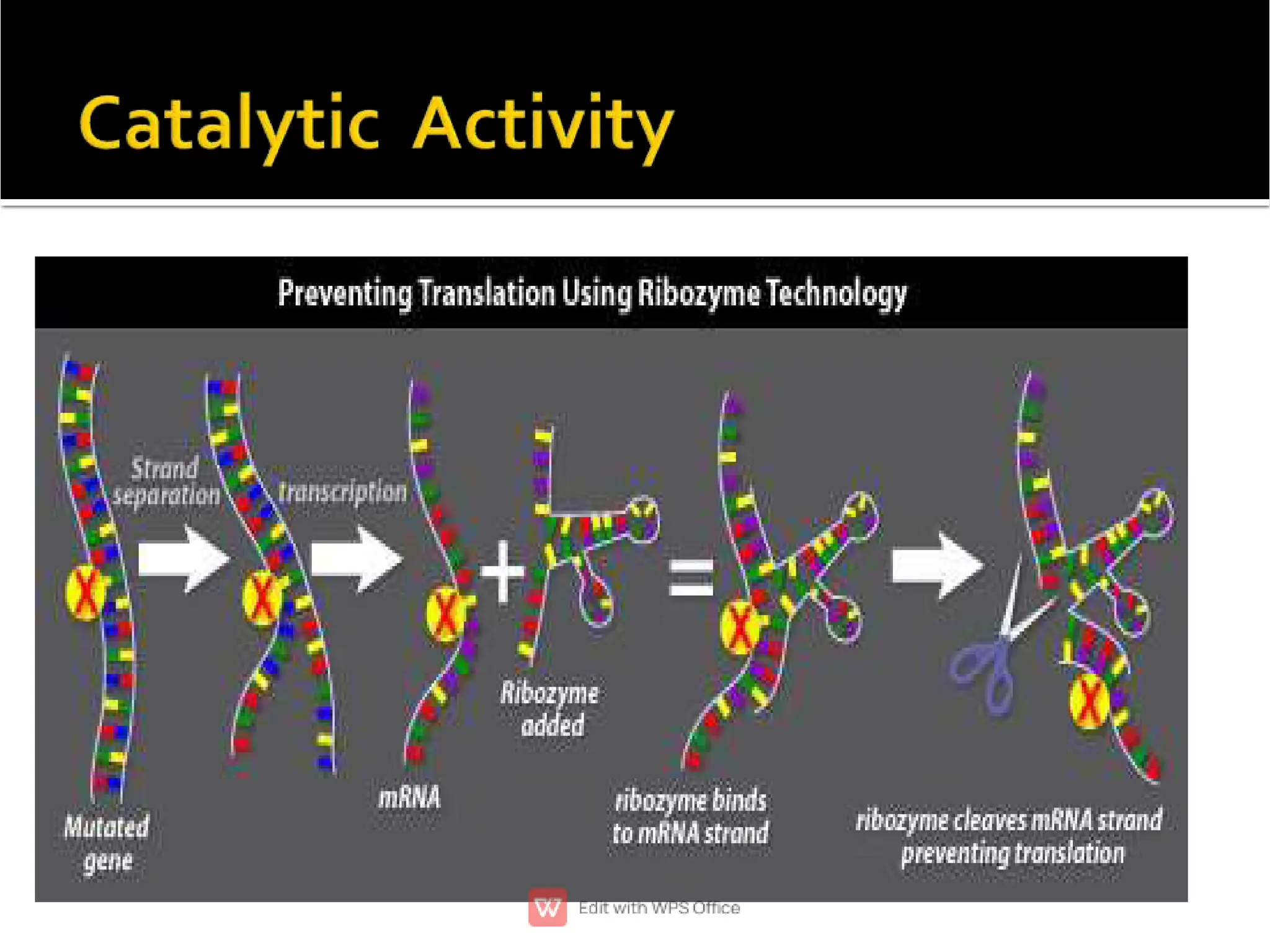
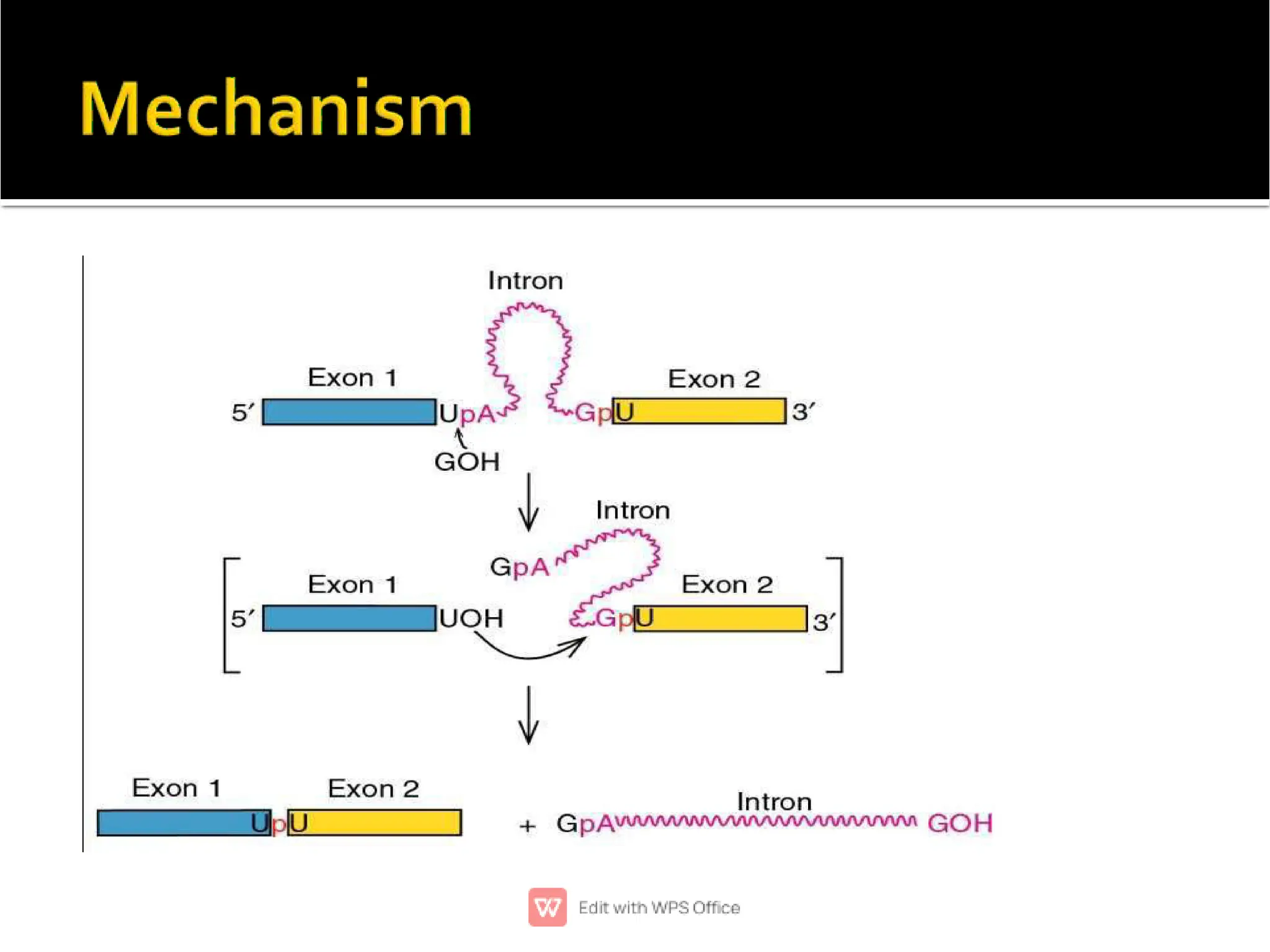
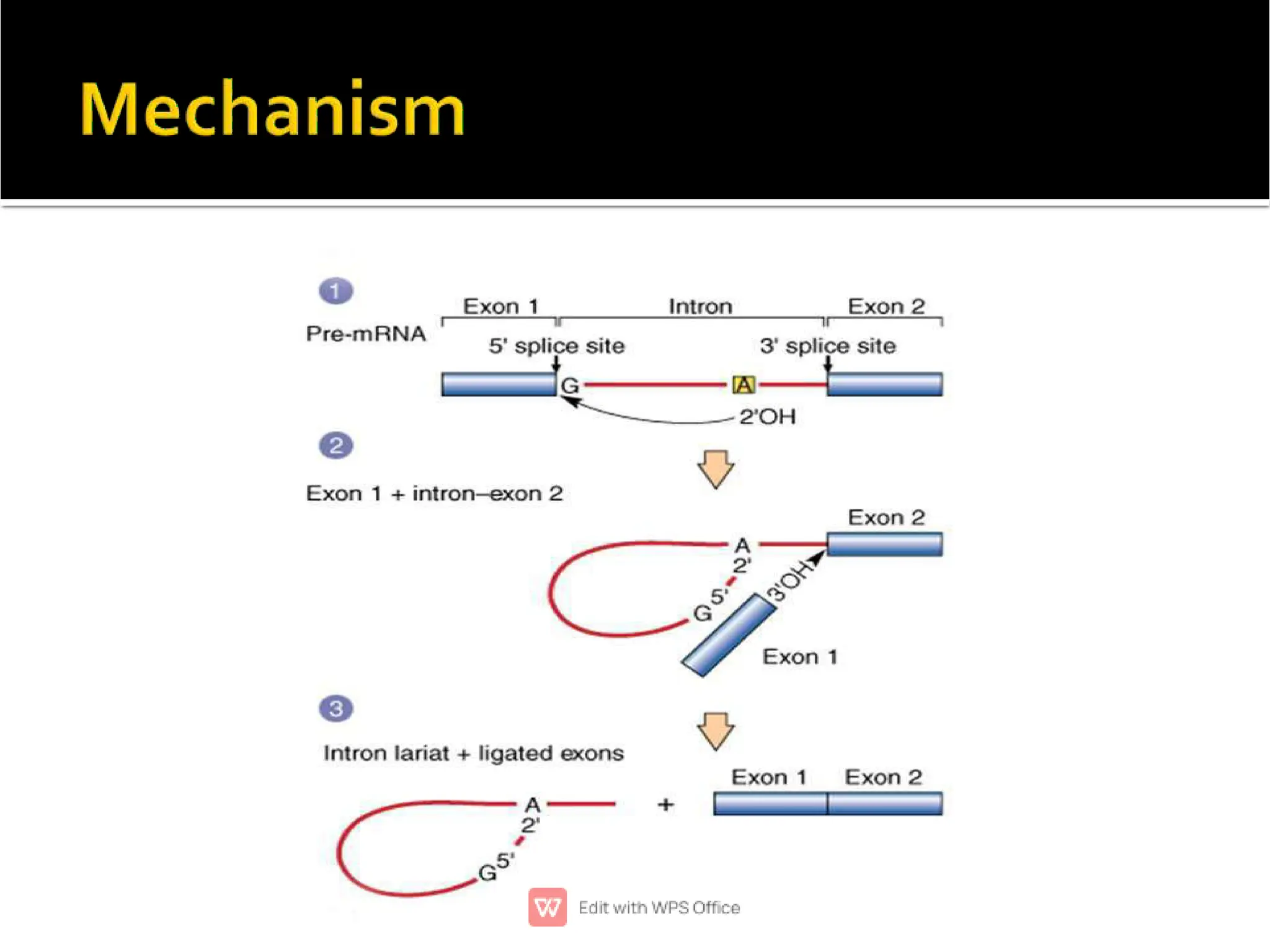
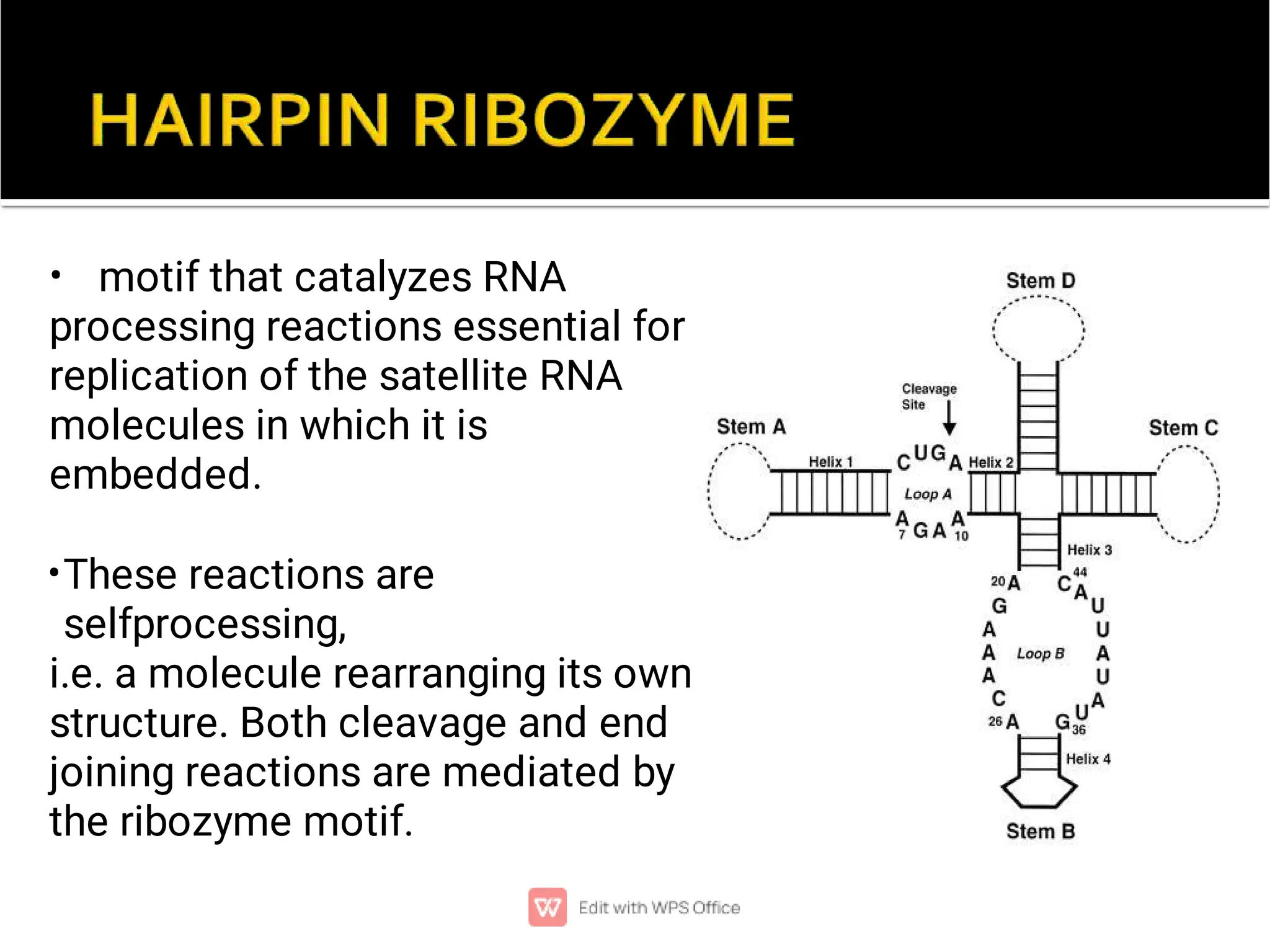
This document provides an overview of split genes and ribozymes. It discusses how split genes have interrupted sequences called introns between exons, and how this was discovered in 1977. It also defines ribozymes as RNA molecules with catalytic activity, describes several natural ribozymes including group I and II introns and hammerhead ribozymes. Applications of engineered ribozymes in gene therapy and inhibiting gene expression are also mentioned.