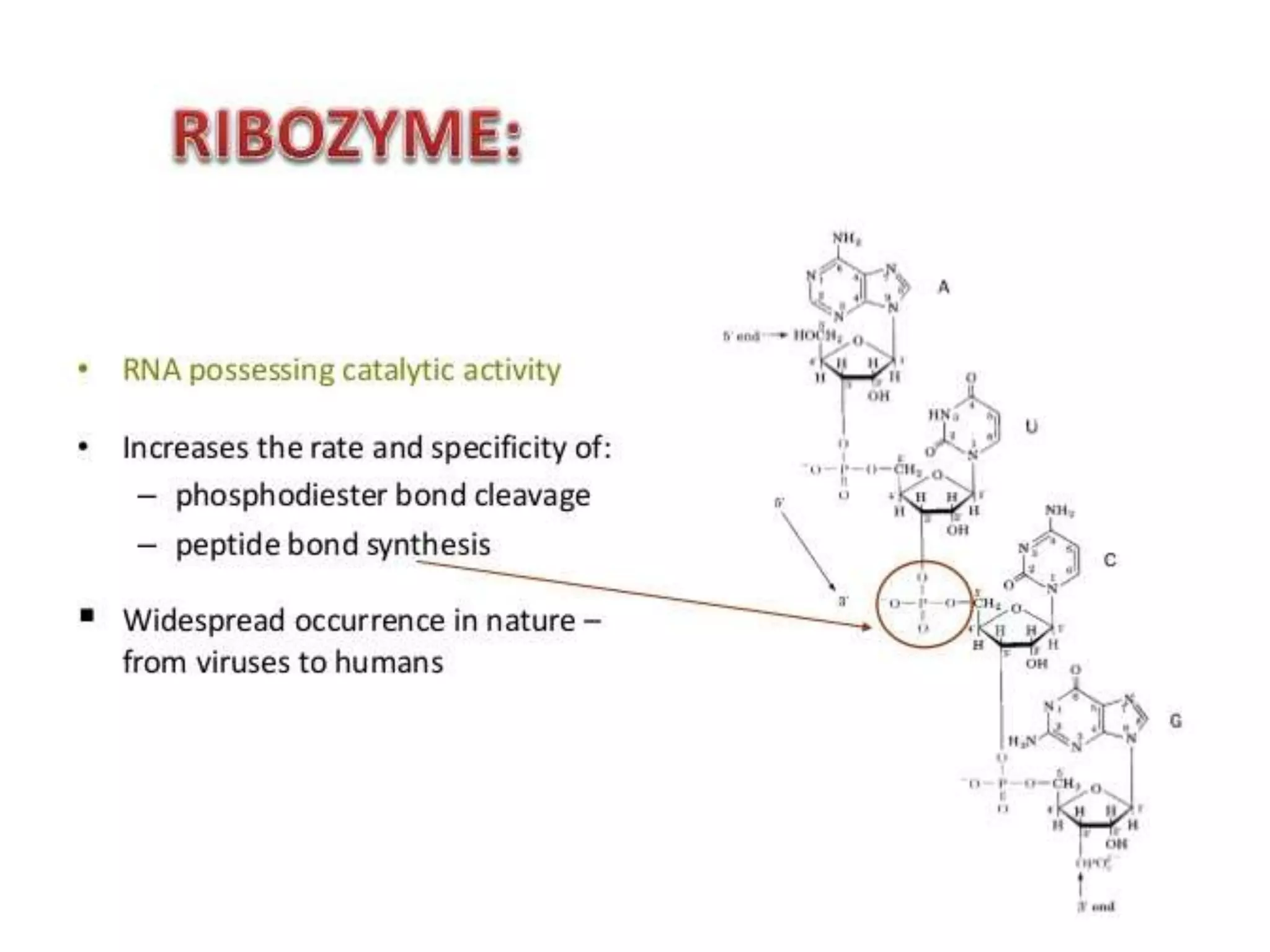
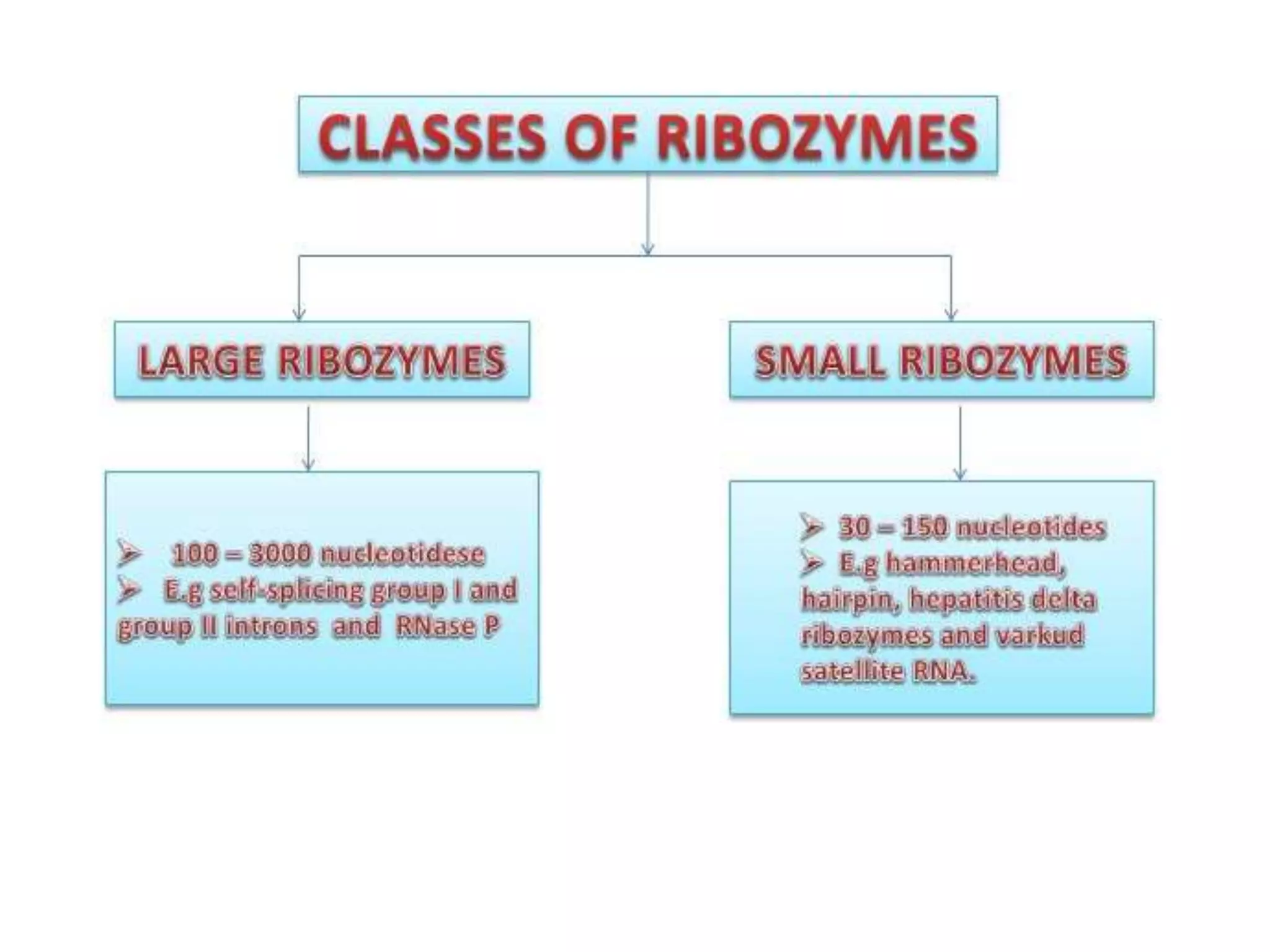
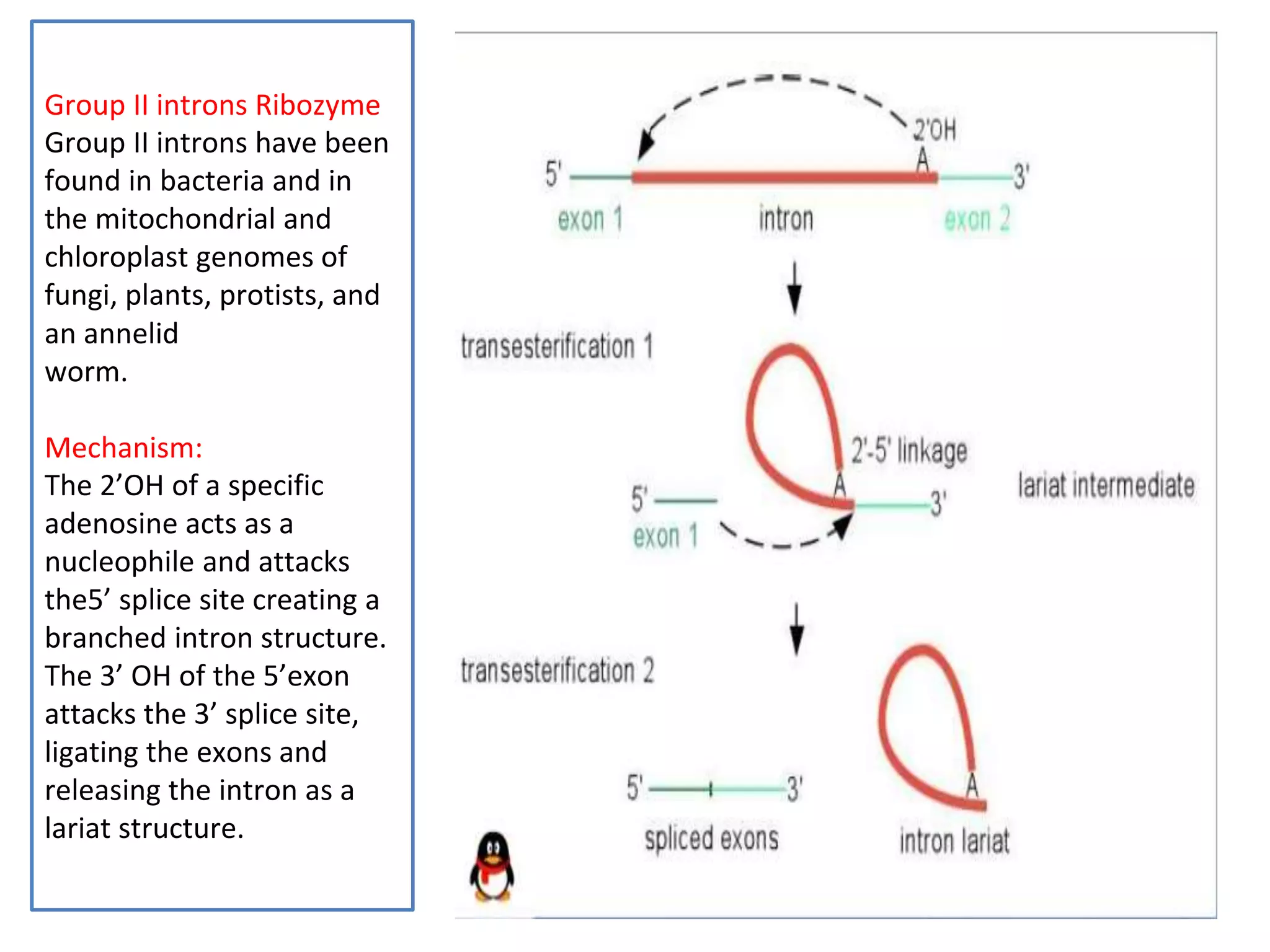
This document provides information about the course Enzymology II including the course code, credits, presenter, and contents. The contents section outlines topics that will be covered such as regulatory enzymes, covalently modified enzymes, catalytic mechanisms of enzymes, specific enzyme mechanisms, isoenzymes, membrane bound enzymes, novel enzymes including ribozymes, cofactors and coenzymes. The document then focuses on ribozymes, describing their characteristics and functions. It discusses naturally occurring ribozymes such as self-splicing introns and artificial ribozymes created in laboratories. Finally, it examines the mechanisms of group I intron and group II intron ribozymes in detail.