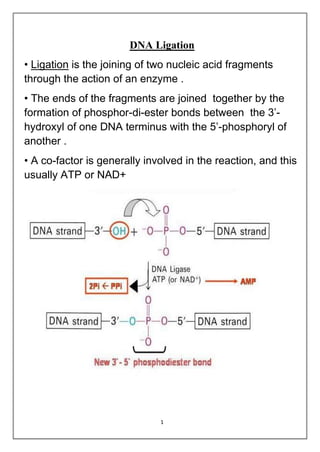
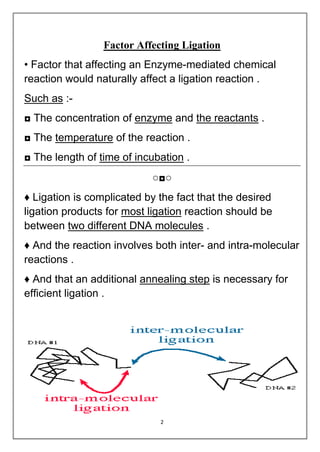
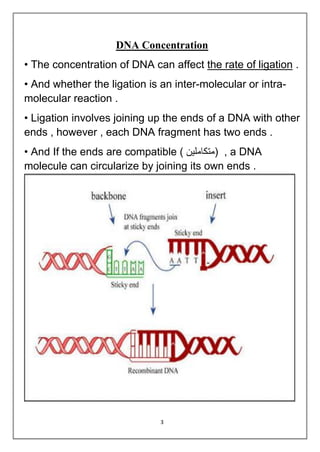
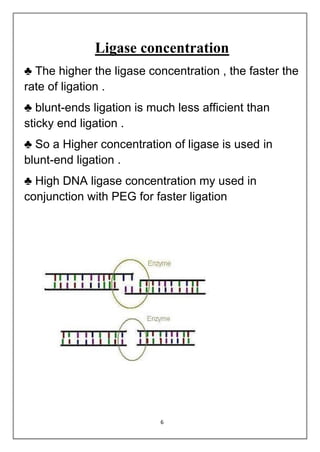
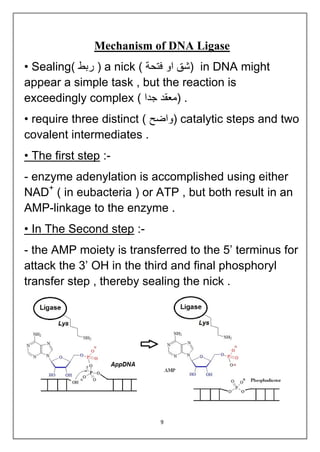
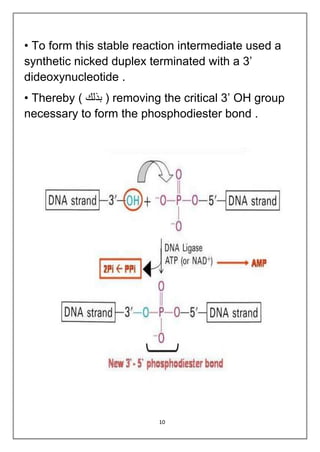
DNA ligation is the joining of two nucleic acid fragments through the action of an enzyme. Several factors can affect the ligation reaction, including the concentration of enzyme, DNA, and cofactors like ATP or NAD+. The DNA concentration is particularly important, as higher concentrations favor intermolecular ligation between separate DNA molecules, while lower concentrations favor intramolecular ligation where a DNA molecule joins its own ends. DNA ligase carries out ligation through a three-step catalytic mechanism involving adenylation of the enzyme and two phosphoryl transfers.