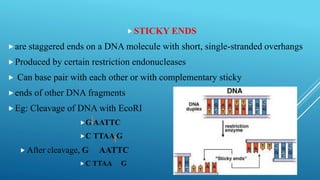
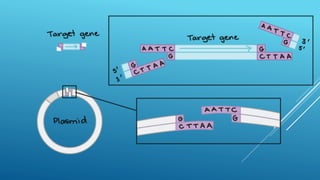
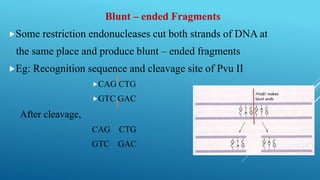
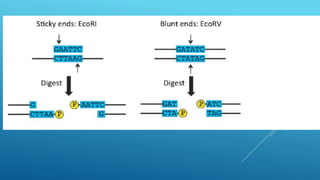
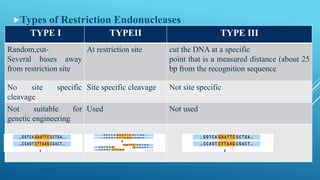
Restriction endonucleases are enzymes produced by bacteria that recognize specific sequences in foreign DNA and cut the DNA at or near the recognition sites. They function as part of the bacterial immune system to degrade foreign DNA. Restriction endonucleases cut DNA into fragments that have either blunt or sticky ends, depending on where they cut the DNA strands. They are named based on the bacteria that produces them and are classified into three types depending on their cleavage patterns. Type II restriction endonucleases cut directly at their recognition sites and produce fragments with sticky or blunt ends, making them useful for genetic engineering applications.