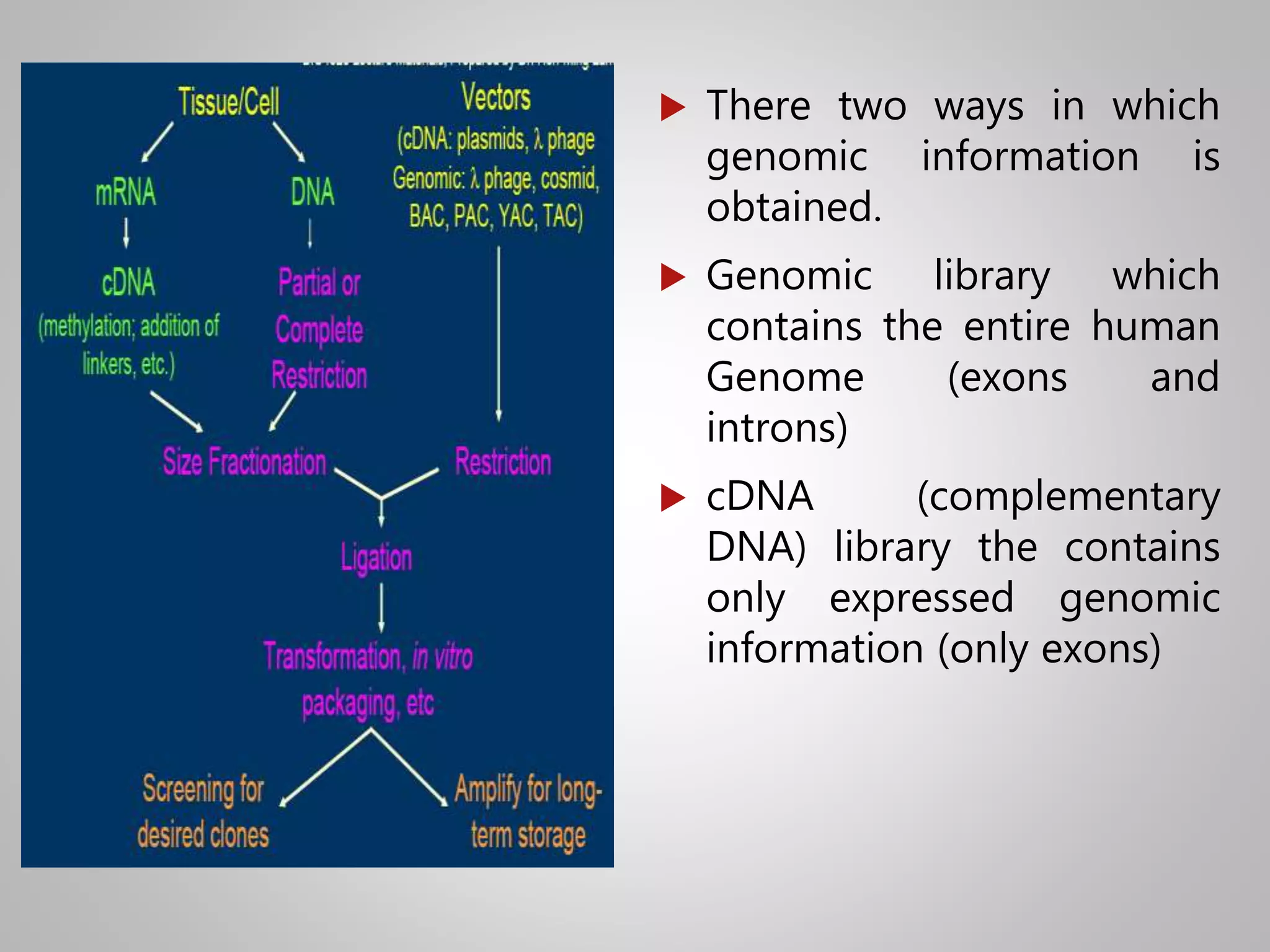
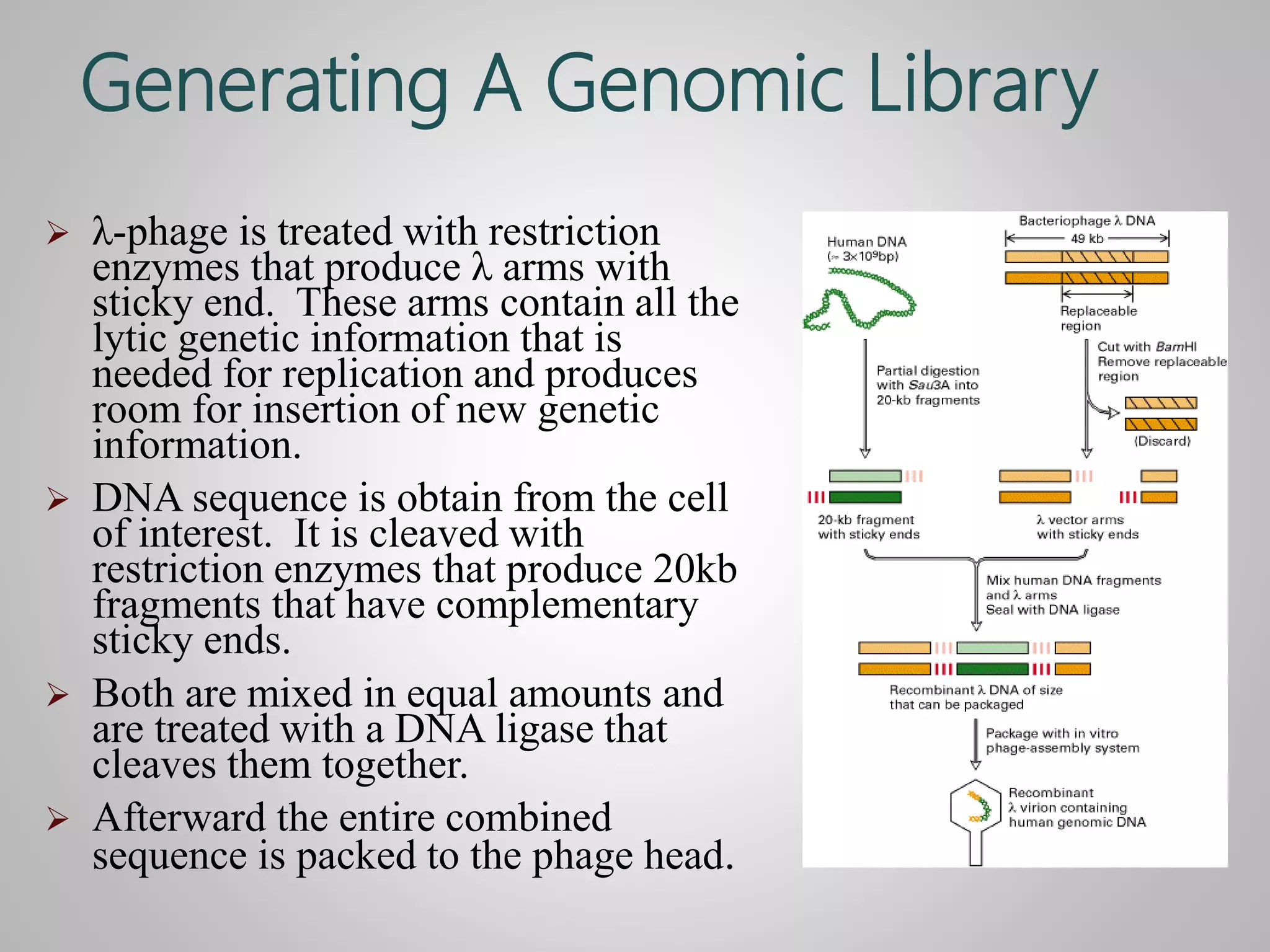
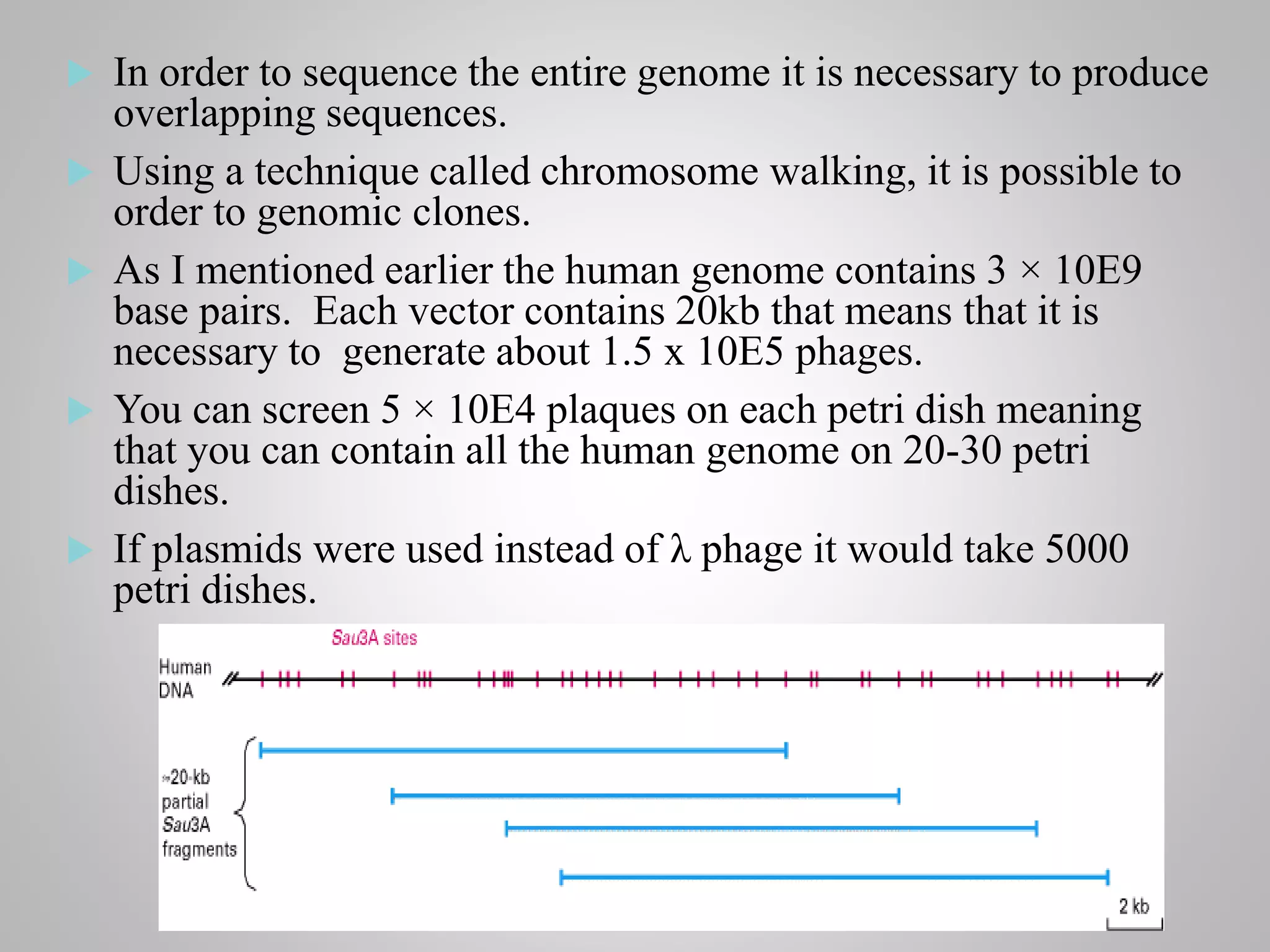
Genomic DNA and cDNA libraries are two ways to obtain genomic information. [1] A genomic DNA library contains the entire human genome, including exons and introns. [2] A cDNA library contains only the expressed portions of the genome (exons). [3] λ-phage is commonly used as the vector to generate these libraries due to its ability to clone large DNA fragments and be packaged in large numbers.