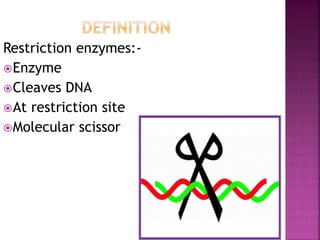
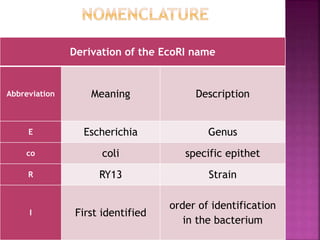
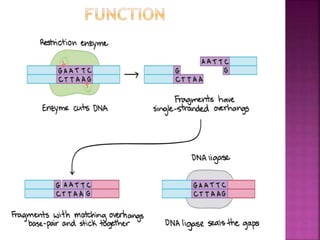
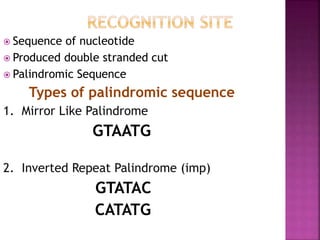
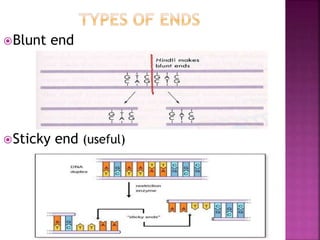
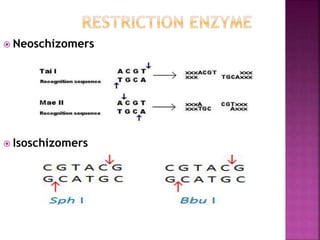
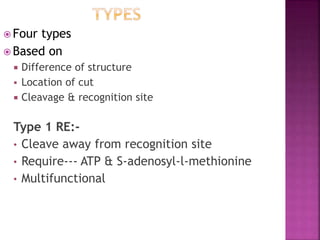
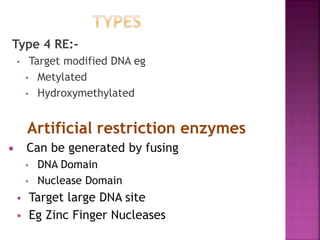
This document provides information about restriction enzymes including their definition, history, origin, types, and applications. Restriction enzymes are enzymes that cleave DNA at specific restriction sites. They were first discovered in the 1960s by scientists studying E. coli bacteria. There are four main types of restriction enzymes that are classified based on differences in their structure and cleavage/recognition sites. Restriction enzymes have various applications in gene cloning, protein expression experiments, and biotechnology.