1) DNA replication begins with the unwinding of the DNA double helix at an origin of replication site.
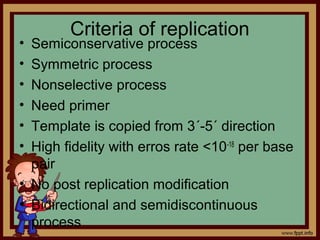
2) This forms a replication fork with leading and lagging strands that are copied semi-conservatively to produce two identical copies of DNA.
3) RNA primers, DNA polymerases, helicase and single-strand binding proteins work together to separate the strands and synthesize new DNA in the 5’-3’ direction along the template.