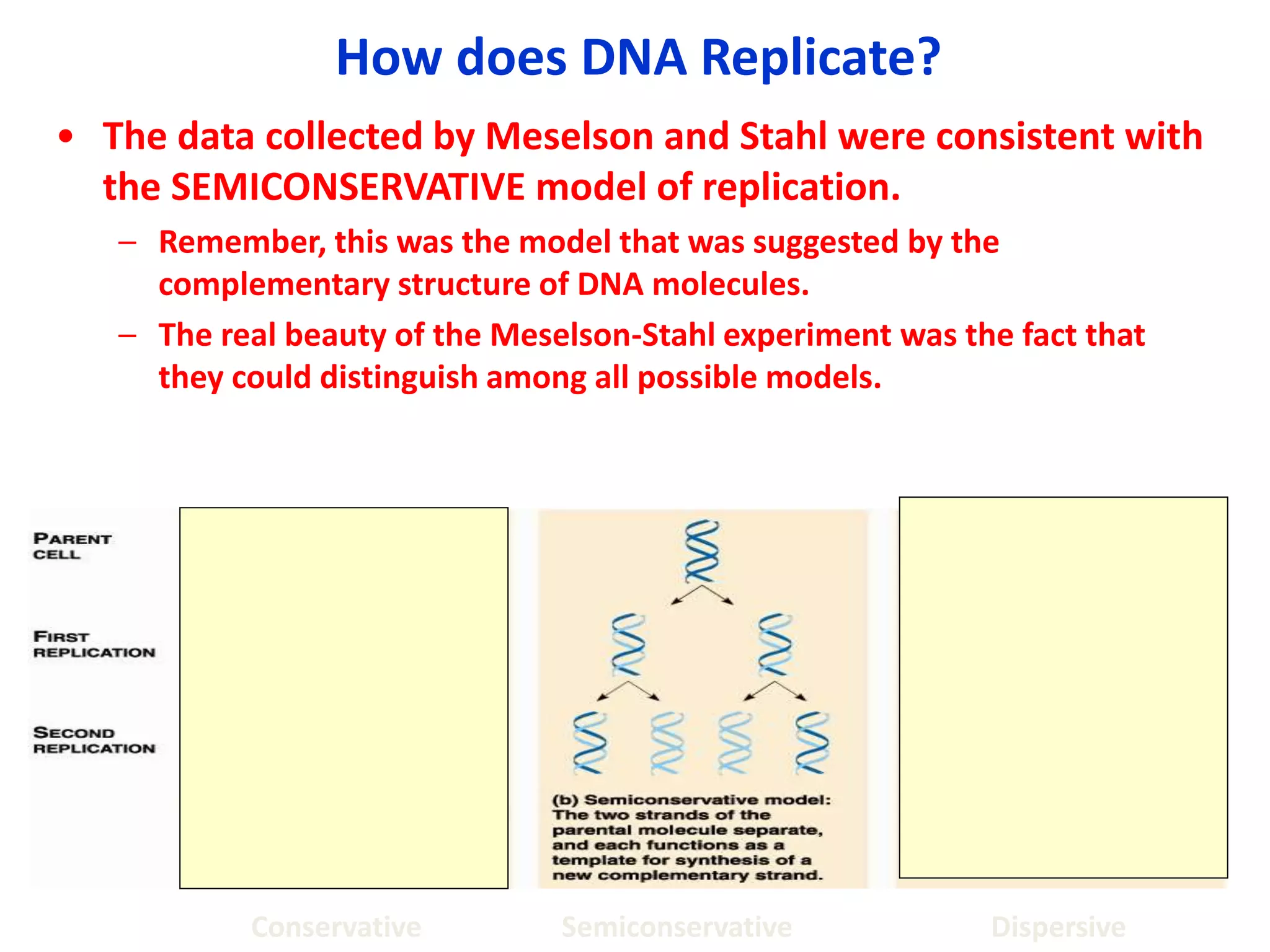
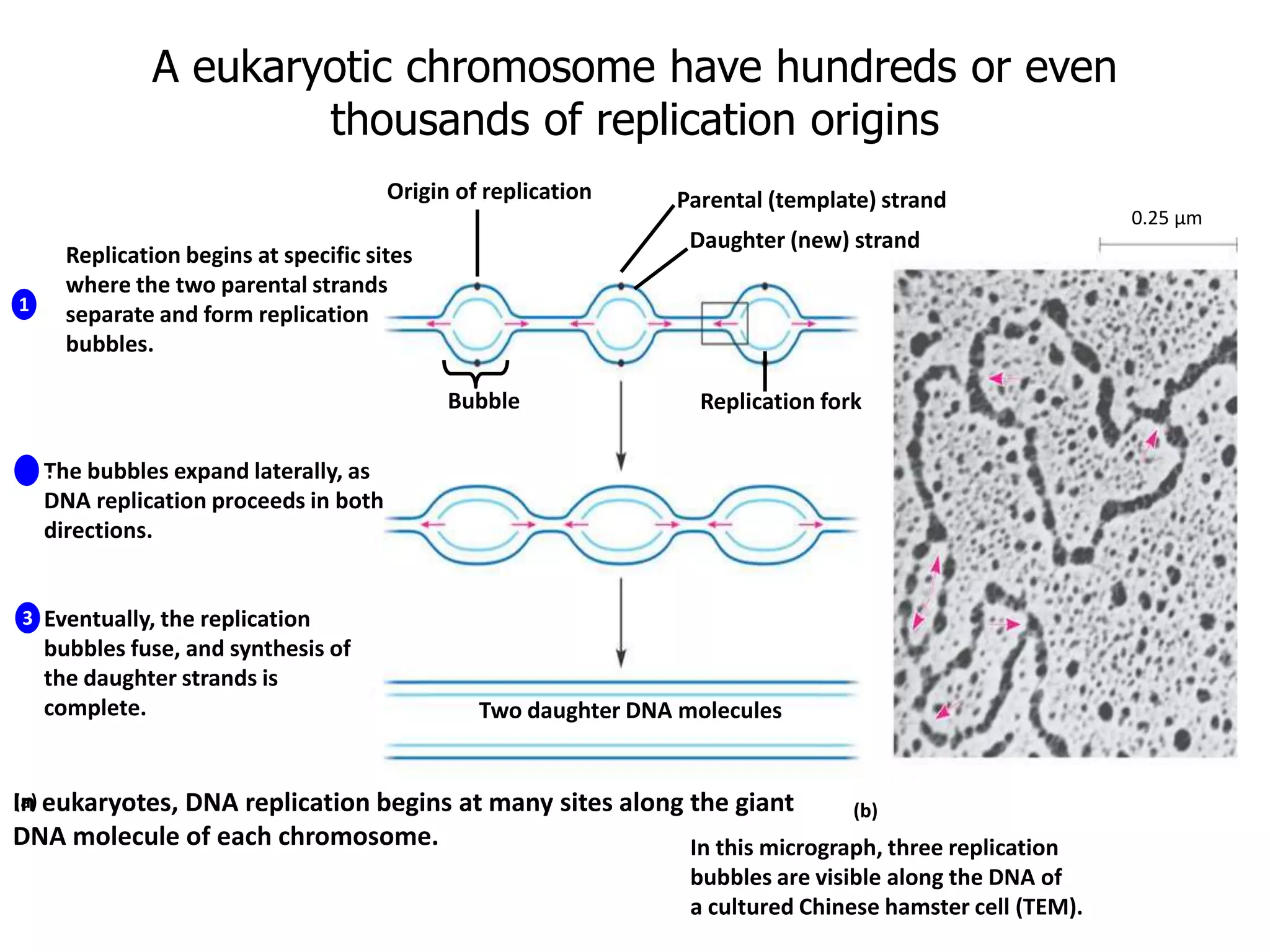
DNA replication involves unwinding the double helix at the replication fork, synthesizing new strands in the 5' to 3' direction, and joining fragments together. It is semi-conservative, with each parental strand serving as a template for a new complementary daughter strand. The leading strand copies continuously from the replication origin, while the lagging strand copies in short fragments that are later joined by DNA ligase. Various enzymes work together in this process, including helicase, gyrase, primase, DNA polymerase, and ligase to make an exact copy of the DNA molecule.