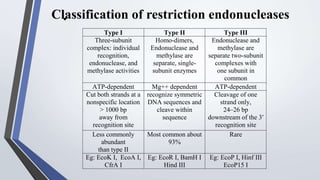
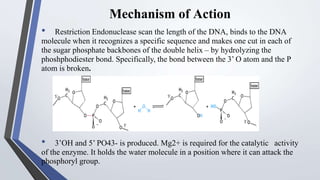
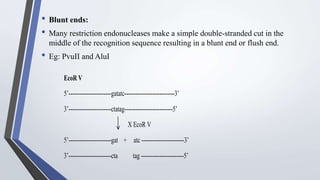
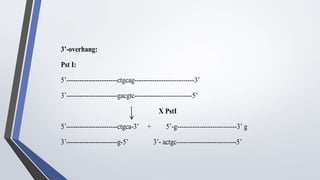
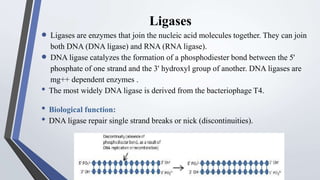
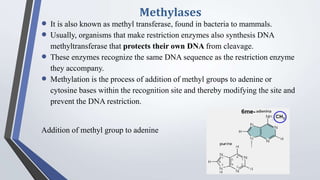
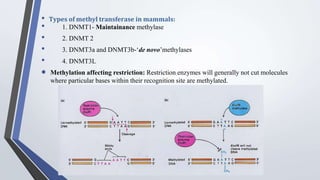
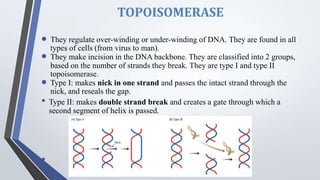
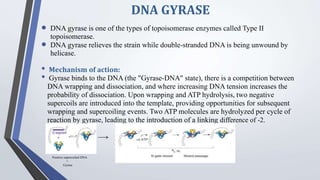
Recombinant enzymes are used in recombinant DNA technology and include nucleases, ligases, polymerases, and DNA modifying enzymes. Restriction enzymes cut DNA at specific recognition sequences and can produce blunt or sticky ends. Ligases join DNA fragments back together. Methylases protect host DNA from restriction enzymes by adding methyl groups to recognition sites. Topoisomerases regulate DNA supercoiling through transient single or double strand breaks. DNA gyrase is a type II topoisomerase that introduces negative supercoils to relieve strain on unwinding DNA.