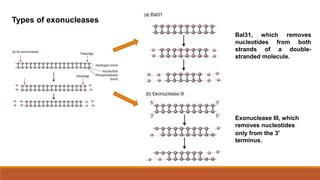
DNA manipulating enzymes include nucleases, ligases, polymerases and modifying enzymes. Restriction endonucleases are enzymes that cut DNA molecules at specific recognition sequences. They produce either blunt or sticky ends. Restriction endonucleases are classified as type I, II or III, with type II having widespread use in recombinant DNA technology by allowing precise cutting of DNA molecules at specific sites.